Pestalotiopsis eleutherococci S. Q. Tian, R. Xu & Phukhams., 2022
|
publication ID |
https://doi.org/ 10.11646/phytotaxa.566.1.6 |
|
DOI |
https://doi.org/10.5281/zenodo.7108280 |
|
persistent identifier |
https://treatment.plazi.org/id/005A4049-FFA9-FFF0-FF64-0DE971A90FC7 |
|
treatment provided by |
Plazi |
|
scientific name |
Pestalotiopsis eleutherococci S. Q. Tian, R. Xu & Phukhams. |
| status |
sp. nov. |
Pestalotiopsis eleutherococci S. Q. Tian, R. Xu & Phukhams. , sp. nov. FIGURE. 2 View FIGURE 2
Index Fungorum number: —IF559721
Etymology: —Name refers the host, Eleutherococcus brachypus
Holotype: — HMJAU 60189
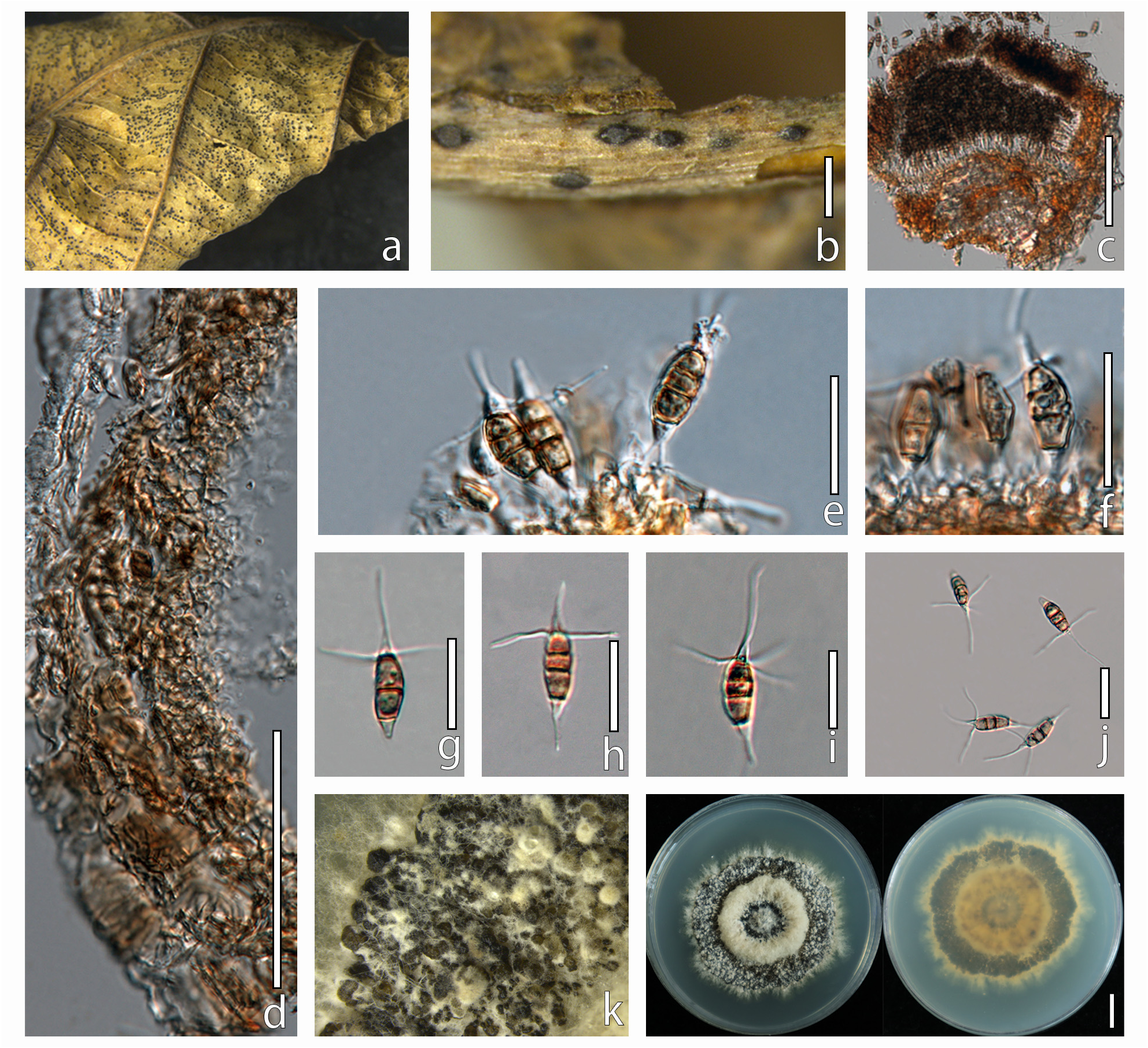
Saprobic on dead leaves of Eleutherococcus brachypus . Sexual morph: Undetermined. Asexual morph: Conidiomata 130–150 × 190–280 μm (x̄ = 140 × 235 μm), pycnidia, sub - globose, depressed, solitary or aggregated, erumpent to semi - immersed, brown to dark brown. Ostioles 36 µm long, 12 µm width, conical, central, brown, spore discharge from the slit-like ostioles of the pycnidia. Conidiophores indistinct, often reduced to conidiogenous cells. Conidiogenous cells 5–20 × 3–7 μm (x̄ = 16.3 × 4.6 μm), apex 2–5 μm diam, discrete, lageniform to subcylindrical, collarette flared, hyaline, smooth, proliferating once or twice. Conidia (21–)22–27(–28) × (5–)5.5–6.5(–7) μm, (x̄ ± SD = 24.7 ± 1.8 × 6 ± 0.6 μm), fusoid, ellipsoid to subcylindrical, straight to slightly curved, 3–4 septate; Two or three median cells (11–)13–18(–19) μm long, (x̄ ± SD = 15.7 ± 1.8 μm), (second cell from the base 3–6 μm long; third cell 3–4 μm long; fourth cell 5–7 μm long), doliiform to subcylindrical, wall rugose, concolourous, golden brown, septa darker than the rest of the cell; Apical cell 4–6.5 μm long, obconic with an acute apex, hyaline, thin and smooth walled, with 2–4 tubular appendage, the median appendage 6–21 μm long, apical appendage 10–19 μm long, attenuated, arising just above the septum of the apical cell and upper median cell, unbranched, hyaline. Basal cell 3–5μm long, narrowly obconic with a truncate base, hyaline, thin and smooth-walled, one basal appendage, 4–8 μm long, single, centric, tubular, hyaline, unbranched.
Culture characteristics: —Colonies on PDA reaching 60–70 mm diam after seven days at 25˚C, sporulating after two weeks of incubation. Cultures from above, dense, round, umbonate, papillate, fluffy, grey with white aerial mycelium; reverse brownish, radiating white to the edge, cream at the margin.
Material examined: — CHINA, Jilin Province: Changchun, from leaves of Eleutherococcus brachypus (Araliaceae) , 18 April 2021, S.Q. Tian and C. Phukhamsakda, T 02 ( HMJAU 60189, holotype); ex - type living culture CCMJ5009, isotype = HMJAU 60190; ex - isotype living culture CCMJ5010.
Notes: —The multi - gene phylogenetic analysis of combined ITS, tef 1 - α, and tub 2 sequence data showed that Pestalotiopsis eleutherococci (HMJAU 60189) is closely related to Pestalotiopsis lijiangensis with strong bootstrap support (ML/MP/BPP: 96%/97%/1.00). In a BLAST search in GenBank, the closest match of the ITS sequence of P. eleutherococci (HMJAU 60189) was 98.28% similar across 99% of the query sequence to P. lijiangensis (CNUZ118), which translates into 97.29% similarity. There was 1.92% (11/573) base differences in the ITS region of P. eleutherococci (HMJAU 60189) and P. lijiangensis (CNUZ118), excluding gaps. The closest match of the tub 2 sequence of P. eleutherococci was 89.21% similar across 93% of the query sequence to P. dianellae (CBS 143421), which translates into 82.07% similarity. There were 6.12% (35/571) and 12.17% (57/468) base differences in the ITS and tub 2 genes of P. eleutherococci (HMJAU 60189) and P. dianellae (CBS 143421), excluding gaps. Morphologically, P. eleutherococci is similar to P. pallidotheae , a species described from Japan, but P. pallidotheae has more (2–5 vs. 2–4) and longer apical appendages than P. eleutherococci (12– 39 μm vs. 10–19 µm) ( Watanabe et al. 2010). Pestalotiopsis eleutherococci (HMJAU 60189) is morphologically similar to P. lijiangensis in the shape of conidia ( Crous et al. 2017, Zhou et al. 2018). However, the appendages (10–19 µm vs. 14–25 µm in P. lijiangensis ) and median cells of P. eleutherococci are smaller (second cell from the base 3–6 μm vs. 4.5–5.5 µm; third cell 3–4 μm vs. 4–5.1 µm; fourth cell 5–7 μm vs. 6–7.4 µm). The basal appendage is longer in P. lijiangensis (10–19 μm vs. 14–25 µm, 4–8 μm vs. 2–6 µm) and the conidial length/ width ratio of P. eleutherococci is almost 1.38 times larger than of P. lijiangensis (4.1 vs 2.96 in the conidial length/width ratio). The third cell of Pestalotiopsis eleutherococci , P. lijiangensis and P. montellica are 0.6–0.78 times compare to the other cells, while the third cell of P. jesteri is 1.1 times larger than the other cells in the conidia ( Strobel et al. 2000). Pestalotiopsis jesteri also has knobbed apical appendages, while P. eleutherococci , P. montellica and P. lijiangensis have unbranched apical appendage. A comparison of the ITS sequences shows that P. eleutherococci (HMJAU 60189) differs by 11 nucleotides from P. lijiangensis , 26 nucleotides from P. jesteri (CBS 109350) and 25 nucleotides from P. montellica (MFLUCC 12 - 0279). Pestalotiopsis eleutherococci differs by 54 nucleotides from P. jesteri and P. montellica , excluding gap in the tub 2 region. There were 4.64% (26/560 bases) and 10.62% (54/508) base differences in the ITS and tub 2 genes between P. eleutherococci and P. jesteri , excluding gaps. There were 4.46% (25/560) and 10.4% (54/519) base differences in the ITS and tub 2 genes of P. eleutherococci and P. montellica , excluding gaps. Therefore, Pestalotiopsis eleutherococci is introduced as a novel species.
| C |
University of Copenhagen |
| T |
Tavera, Department of Geology and Geophysics |
| HMJAU |
Herbarium of Mycology of Jilin Agricultural University |
No known copyright restrictions apply. See Agosti, D., Egloff, W., 2009. Taxonomic information exchange and copyright: the Plazi approach. BMC Research Notes 2009, 2:53 for further explanation.
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |