Lordomyrma sinensis
|
publication ID |
https://doi.org/ 10.5281/zenodo.190065 |
|
publication LSID |
lsid:zoobank.org:pub:AFB7BDC6-2973-482F-BEB5-4878BCBFA4B3 |
|
DOI |
https://doi.org/10.5281/zenodo.5682102 |
|
persistent identifier |
https://treatment.plazi.org/id/03A0D36A-114C-9D05-FF3F-6176171AFE68 |
|
treatment provided by |
Plazi |
|
scientific name |
Lordomyrma sinensis |
| status |
|
Lordomyrma sinensis (Ma, Xu, Makio, and DuBois) comb. n.
Stenamma sinensis Ma, Xu, Makio , and DuBois, 2007: 371–377, Figs. 1–4 View FIGURE 1 View FIGURES 2 – 16 . Holotype worker and paratype workers, CHINA: Mt. Qinling, Shaanxi, 33º39`N 107º48`E, 1580–1641m, 7–18 September 2005 and 1–13 August 2006, Li- Bin Ma. [Holotype and partype workers in DBSNU not examined.]
Justification for transfer of species to Lordomyrma
Morphological analysis
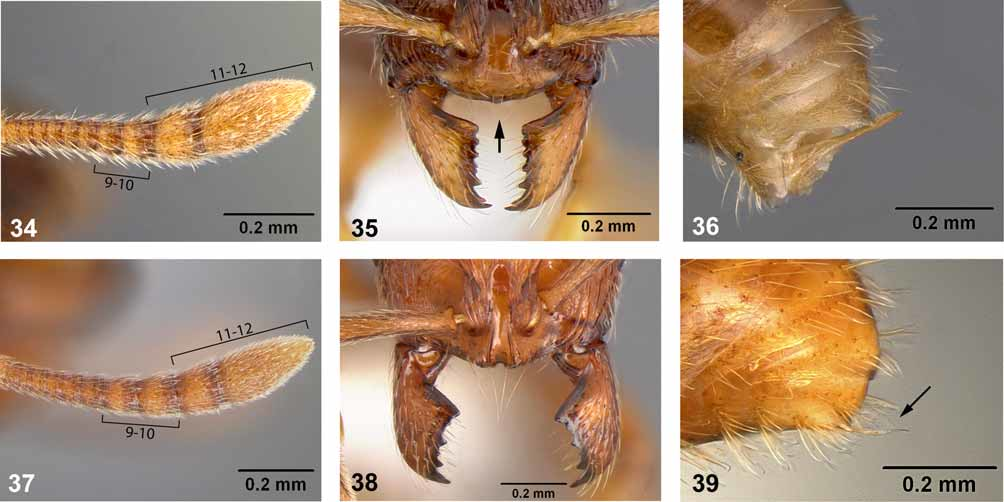
The holotype and several paratype specimens of Lordomyrma bhutanensis from the NHMB were examined. Additional material recently collected from Nepal and Yunnan Province, China was borrowed from MCZC and CASC, respectively. These latter specimens were sorted into two morphospecies and identified as L. cf. bhutanensis 1 and L. cf. bhutanensis 2 ( Figures 28–33 View FIGURES 25 – 33 ). Careful examination revealed several morphological characters distinguishing these species from Stenamma : (1) Antenna with a 3-segmented club of which the last two segments display the largest increases in length relative to preceding segments (ACI 73- 77; compare Figures 34 and 37 View FIGURES 34 – 39 ); (2) apex of anterior clypeal margin with a small projecting tooth (compare Figures 35 and 38 View FIGURES 34 – 39 ; note, this character was not discerned in the types of L. bhutanensis because the mandibles were closed in all specimens); (3) posteromedial margin of clypeus as wide or wider than frontal lobes in fullface view (compare Figures 35, 38 View FIGURES 34 – 39 ); (3) postpetiolar node broader than long ( Figures 27, 30, 33 View FIGURES 25 – 33 ); (4) sting robust and often exposed and projecting dorsally in pinned specimens (compare Figures 36 and 39 View FIGURES 34 – 39 ). Although specimens of L. sinensis were not examined, the characters mentioned above were confirmed by reviewing the species description and accompanying figures ( Ma et al. 2007).
To adequately delimit species in this group, a more detailed analysis of regional variation will be necessary. From the specimens examined here, I find it difficult to comfortably define species boundaries. Surprisingly, Ma et al. (2007) did not directly compare L. sinensis to L. bhutanensis , but instead stated that it would most likely be confused with other species within the Stenamma owstoni species group, the group to which both L. sinensis and L. bhutanensis were assigned. I examined three additional members of this species group, S. koreanensis , S. owstoni , and S. nipponense ( Figures 5–7 View FIGURES 2 – 16 ), and determined that these have the attributes of true Stenamma . It is likely that this confusion was caused by a poor understanding of which characters are most important in diagnosing Stenamma . For example, Ma et al. (2007) were the first to notice the presence of a median clypeal tooth. However, they incorrectly used this trait as a species diagnosing rather than a genus diagnosing character.
Phylogenetic methods
The DNA from one specimen each of L. cf. bhutanensis 1 and L. cf. bhutanensis 2 was non-destructively extracted. Both specimens were collected from sifted leaf litter in forest, Shibali, Yunnan Province, China at 2,475 m. Shortly after extraction, I obtained fragments of four nuclear genes from both taxa: 28S rDNA, abdominal-A (abdA), elongation factor 1-alpha F2 copy (EF1αF2), and long-wavelength rhodopsin (LW Rh). DNA extraction, amplification, and sequencing were performed as in Brady et al. (2006). All newly generated sequences have been uploaded into GenBank (see Table 1 View TABLE 1 for specimen codes and GenBank accession numbers). Preliminary phylogenetic analyses were then performed using both a comprehensive Stenamma data set, which included over 30 species representing all regions where Stenamma is found (Branstetter unpubl.), and a large myrmicine data set containing 63 myrmicine genera and several outgroups (Ward pers. com.) Both data sets revealed that L. cf. bhutanensis 1 and L. cf. bhutanensis 2 fall outside of the genus Stenamma and the myrmicine data showed consistent association between L. cf. bhutanensis and the genus Lordomyrma .
Following these initial results, a more rigorous analysis was performed. The above gene fragments were obtained from four additional Stenamma species ( Stenamma expolitum , S. felixi , S. meridionale , S. striatulum ), Cyphoidris exalta Bolton (Ward pers. com.), Lordomyrma desupra Sarnat (Ward pers. com.), and L. epinotalis Mann (Lucky and Sarnat pers. com.). Cyphoidris exalta was included in these analyses because it has been hypothesized to be the sister group to Lordomyrma ( Bolton 1981; Taylor 2009). These sequences were then incorporated into the 162-taxon Ant Tree of Life data set of Brady et al. (2006). This data matrix was culled to include the majority of myrmicine species and several outgroups for a final 63-taxon data matrix. Sequence alignment was performed using default settings in the program ClustalX v1.83.1 ( Jeanmougin et al. 1998) and manually edited with MacClade v4.08 ( Maddison & Maddison 2005). An intron in LW Rh and hypervariable regions of 28S were excluded from all analyses. The final data matrix included 2326 base pairs, with 646 parsimony informative sites, and 826 variable sites.
Taxon Country Specimen Code Voucher Type Deposited LW Rh EF1aF2 abdA 28s Phylogenetic analyses employed Bayesian and maximum likelihood (ML) methods and were run through the Cyberinfrastructure for Phylogenetic Research (CIPRES) computer cluster at the San Diego Super Computer Center (http://www.phylo.org/). Four partitioning schemes were employed: single partition, partitioning by gene, partitioning by codon position (positions 1+2 versus 3) with 28S forming its own partition, and partitioning by gene and codon position. This resulted in a one-, four-, three-, and sevenpartition model, respectively. The model of sequence evolution for each partition was selected using the AIC with MrModeltest v2 ( Nylander 2004b; Posada & Crandall 1998). This resulted in the model GTR+I+G being selected for all partitions except LW Rh (HKY+I+G) and EF1αF2 codon positions 1+2 (SYM+I+G). Bayesian analyses were conducted using MrBayes v3.1.2 ( Ronquist & Huelsenbeck 2003) under each of the four partitioning schemes mentioned above. For each analysis, two independent MCMC runs were performed for 5 million generations, distributed across four chains with the default heating parameter. Convergence between runs was assessed using the average standard deviation of split frequencies and by plotting likelihood values across generations using Tracer v1.4.1 ( Rambaut & Drummond 2007). A burn-in value of 500,000 generations was established and only the post-burn-in generations from both runs were included in the results. Bayes factor comparisons ( Nylander 2004a) showed that the best partitioning scheme was by gene and codon position. Maximum Likelihood inference was implemented in the rapid bootstrapping program RAxML ( Stamatakis et al. 2008). First, single gene analyses were performed to look for aberrant results. Next, the combined data set, partitioned by gene and codon position, was analyzed and RAxML was set to find the highest scoring ML tree and to perform 1000 ML bootstrap replicates. The GTR+CAT model of DNA sequence evolution was applied to all partitions in RAxML.
No known copyright restrictions apply. See Agosti, D., Egloff, W., 2009. Taxonomic information exchange and copyright: the Plazi approach. BMC Research Notes 2009, 2:53 for further explanation.