Locustella portenta, Rheindt & Prawiradilaga & Ashar & Lee & Wu & Ng, 2020
|
publication ID |
https://doi.org/10.5281/zenodo.3608758 |
|
publication LSID |
lsid:zoobank.org:pub:8114B399-C68D-43C2-B6D3-B51AA898431E |
|
DOI |
https://doi.org/10.5281/zenodo.3610533 |
|
persistent identifier |
https://treatment.plazi.org/id/2A588C42-4DBE-41A3-A95A-7B6850521944 |
|
taxon LSID |
lsid:zoobank.org:act:2A588C42-4DBE-41A3-A95A-7B6850521944 |
|
treatment provided by |
Plazi |
|
scientific name |
Locustella portenta |
| status |
sp. nov. |
SM3:
Locustella portenta , species nova
( Taliabu Grasshopper-Warbler;
urn:lsid:zoobank.org:act:2A588C42-4DBE-41A-A95A-7B6850521944
) Frank E. Rheindt, Dewi M. Prawiradilaga, Hidayat Ashari, Suparno, Chyi Yin Gwee, Geraldine W. X. Lee
Holotype
MZB.Ornit.34.411 ( fig. S3 View Fig ); adult female collected 12 Dec 2013 near Waiyo dinahana Camp (~ 1200m) on Taliabu Island ( S 01⁰ 47.614 '; E 124⁰ 48.216 '). Collected by the Rheindt / LIPI field party, including tissue samples from breast muscle and liver; skin prepared by Suparno; field number Tbu80; heavy body molt with fresh remiges; medium fat; weight 19g; wing length 5.9cm; wing spread 18.4cm; total length 13.9cm; bill 1.4cm; tail 5.4cm; tarsus 2.5cm.
GoogleMapsDescription of holotype
The entire crown from forehead to nape is rich earthen-brown (7.5YR 3/3) with slightly darker feather margins creating a scalloped appearance. A thin off-white supercilium is pronounced from bill base to just behind eye, from where it slowly tapers out. Lores are dark brownish-black (2.5Y 1.5/1). Ear coverts are of a medium earthen-brown tone (5YR 3/1) with fine pale shaft streaks giving a grizzled appearance, grading into a more greyish-brown tone (10YR 6/1) in the moustachial and malar region. The mantle and scapulars down to the uppertail coverts are largely concolorous with the crown but lack the scalloping created by darker feather margins. The remiges are mostly a rich cold dark-brown (2.5YR 2/1) but have warmer outer webs concolorous with mantle. The upperwing coverts are similarly two-toned, being mostly concolorous with mantle but some showing darker outer webs concolorous with most of remiges. As typical for the genus Locustella , the rectrices are narrow and dark- brown, very close in color to the remiges (2.5YR 2/1). The tail feathers are mostly worn, although one in the centre appears to be a new replacement.
The white chin and throat are quite sharply demarcated from the face. The lower throat and upper breast, still white, are interspersed with fine dusky speckling increasing towards the breast and lower throat sides where the color grades into a solid mid-grey (N5) with only few inconspicuous dusky speckles. The grey breast color then grades into a tawny- ochraceous belly color (7.5YR 4/6), first through tracts of grey feathers which have warm- ochraceous edging, then through tracts of largely ochraceous feathers that still have a grey feather centre, eventually to all-ochraceous feathers. The belly color becomes darker and richer on the undertail coverts and flanks (7.5YR 3/4).
The bill is black with an inconspicuous paler tip. The gape on the live bird was slightly enlarged light yellow. The iris is dark brown. The tarsus and toes were described on the live bird as maroon-horn with paler, more yellowish toepads.
Diagnosis
A rich dark-brown forest warbler typical of the montane resident Locustella radiation from throughout Indonesia and the Philippines. As is usual across this cryptic genus, the new taxon is most conspicuous not on the basis of its plumage but its vocalizations.
Morphologically, it is most similar in size and shape to L. castanea of Sulawesi. However, in comparison to the new taxon, L. castanea has less distinct speckling on the breast, has a more uniform crown that is less scalloped, paler and warmer brown upperparts, greyer underparts with brown mostly only on flanks, and has a less pointed, wider tail.
L. musculus from Seram differs in its less distinct central breast spotting, greyer (less whitish) supercilium, and more extensive grey on breast reaching further onto upper breast, neck sides and auriculars.
L. disturbans from Buru differs in its much whiter breast, with off-white reaching onto mid-belly, whereas only the lower flanks and lower belly are warm-brown. Although the central breast of L. disturbans has a mid-grey tinge, its underparts are much paler than the new species’ fairly uniform grey lower breast, which grades into a fairly dark homogeneous warm-brown belly.
The new species is quite different from the smaller L. montis from Java. The latter is much paler cinnamon-brown on upperparts, crown and especially remiges and rectrices, and is uniformly buff-white below with extensive dark-brown speckling from chin to upper belly, lacking the new species’ grey tones on the breast and dark warm-brown tones on the belly.
Etymology
The name portenta – past participle of Latin ‘portendere’ – roughly translates as “the predicted one” or “the foretold one” and relates to the unusual circumstances of discovery when FER first heard the new warbler and recognized it to be a novel species of Locustella multiple days before a visual confirmation was obtained.
16
Individual, sex and age-related variation within the taxon
Currently only one adult specimen is available (i.e., the holotype), and photos of up to two more adults suggest that this is probably a sexually monomorphic species with limited plumage variation.
History of discovery
FER was the first person to discover L. portenta on 9 April 2009 near its type locality when he heard an unmistakable insect-like sound that he attributed to a hitherto unnamed taxon of Locustella grasshopper-warbler. Having been exposed to the song of L. castanea on a visit to Sulawesi only two weeks prior, and having had previous field experience with L. disturbans and L. musculus from Buru and Seram, respectively ( 8 0), FER was aware that the song of this population – albeit superficially reminiscent of the others – is distinct at the species level. Even so, because of the retiring nature of this bird and ill-timed heavy rain, it took FER another two hiking expeditions to the highlands of Taliabu to finally confirm on 15 April 2009 that this song was indeed uttered by what he perceived as an “unusually dark” individual of grasshopper-warbler ( 48). We encountered this population again during our collecting trip to this area from 6-16 Dec 2013 ( 19) and collected a single specimen, now the holotype.
Distribution and status
L. portenta is presumably restricted to montane forest at the highest elevations of the island of Taliabu ( 19, 48) ( fig. S2 View Fig ). All our records are from elevations above 1,050m on the more mountainous western half of the island of Taliabu, which reaches its highest altitude of ~1,415m at an unnamed peak near our collection locality ( fig. S2 View Fig ). The new species inhabits a peculiar type of dwarf montane forest typical of these elevations.
The occurrence of the new species on neighboring islands is highly doubtful. Of the three major islands that form the Sula archipelago, Taliabu is the largest and tallest. The smallest main island, Sanana, barely reaches above 600m elevation. The highest elevation on its larger neighbor Mangole is 1,127m – theoretically within the elevational range of this grasshopper-warbler – but the area of land above 1,050m on Mangole is only ~36 ha, which is too small an area to allow for the survival of a highly montane bird species throughout evolutionary time scales. On the neighboring Banggai archipelago, the situation is similar: the only island high enough to allow for the theoretical possibility of grasshopper-warbler breeding occurrence is Peleng (highest elevation ~1,022m), but extensive fieldwork around this highest area has not revealed the presence of any resident grasshopper-warblers ( 19, 49).
L. portenta must be a rare bird: Taliabu’s area above 1,050m amounts to ~16,690 ha of formerly forested land in the interior of the island’s western half, over 97% of which forms one contiguous block ( fig. S2 View Fig ). However, satellite imagery predominantly from 2004 indicates that a total of ~1,023 ha (~6.1%) of this area had been affected by logging operations that have degraded the forest and removed its undercover, presumably rendering the affected areas unsuitable, or nearly so, for the grasshopper-warbler. In the one-and-a-half decades since the satellite images were obtained, extensive additional logging is likely to have occurred. Another 940 ha (~5.6%) of the area above 1,050m have succumbed to a series of catastrophic forest fires in 1982 and 1983, and now constitute open grassland habitat unsuitable for this warbler ( fig. S2 View Fig ). Our surveys showed that L. portenta does not occur in the extensive recently generated grasslands in the highlands of Taliabu ( 48). This leaves L. portenta with less than 15,000 ha of suitable habitat, an area likely to further diminish in size with ongoing habitat destruction and the impacts of global warming leading to an upward elevational shift of habitat zones.
Taxonomic rationale
We include this new taxon within Locustella in the family Locustellidae following the phylogenetic demonstration that the genus Bradypterus (previously used for this Southeast Asian clade of warblers) is restricted to Africa ( 8 1, 8 2). Its inclusion within Locustellidae is the reason why we follow the recent trend [e.g. ( 57)] of using the English moniker ‘grasshopper-warbler’ rather than the term ‘bush-warbler’, which is more appropriately reserved for members of a radiation in the family Cettiidae .
The genus Locustella is renowned for its cryptic and conservative coloration that has confounded plumage-based classifications for decades and led to delays in the recognition of species-level taxa until bioacoustic and molecular methods have been applied ( 8 3, 8 4). In Wallacea, members of a Locustella radiation from Sulawesi and the Moluccas were previously united into a single polytypic species L. castanea ( 6 1, 7 5), but have recently been divided into three constituent island species, L. castanea from Sulawesi, L. musculus from Seram, and L. disturbans from Buru, on the basis of their strikingly different songs ( 57). Our new L. portenta is geographically surrounded by constituents of this radiation and is a member of it both on the basis of morphological characters (see Diagnosis) and bioacoustic ones.
Although plumage distinctions are not as important in the genus, L. portenta is characterized by comparatively unique, dark-brown upperparts that stand in contrast to the generally paler and warmer-brown upperparts of neighboring species. However, we mainly attribute species status to L. portenta on the basis of its unique song and genetic differentiation. We here provide an excerpt of ongoing analyses as relating to the bioacoustic and mitogenomic differentiation of L. portenta from neighboring species.
19
Bioacoustic evidence: A total of 41 suitable, high-quality, homologous Locustella song recordings from the field were gathered for analysis from various sources, encompassing all three previous species of the L. castanea radiation plus the new species. These recordings were measured for 11 suitable bioacoustic parameters, and resultant measurements were subjected to principal component analysis (PCA) and a conservative vocal diagnosability criterion ( 85), henceforth referred to as the Isler Criterion, to assess song diagnosability among taxa. Further details regarding all analyzed recordings, such as GPS locations, dates, times, and names of recordists, are listed in table S3, while more details on bioacoustic methodology are provided below under separate Methods paragraphs.
Based on the Isler Criterion, eight out of eleven song parameters measured were diagnosable between the new Taliabu Grasshopper-Warbler L. portenta and two other taxa: L. musculus from Seram and L. castanea from Sulawesi (table S4). Four out of eleven parameters were Isler-diagnosable between the new Taliabu Grasshopper-Warbler and L. disturbans from Buru (table S4). The ‘yardstick approach’ ( 70) can be adopted to ascertain if the level of vocal divergence between these archipelagic taxa is comparable in magnitude to divergences across unequivocal species-level divisions. When comparing Taliabu Grasshopper-Warbler song against that of the Friendly Grasshopper-Warbler L. accentor , a geographically distant species from montane Borneo that has never been included in the L. castanea radiation, only two parameters emerged as Isler-diagnosable (table S4). Using this result as a yardstick for what constitutes species level vocal differentiation, the magnitude of vocal divergence between Taliabu Grasshopper-Warbler and L. musculus on Seram, L. disturbans on Buru, and L. castanea on Sulawesi is extremely high, indicating they all have diagnosably distinct vocalizations that are best reflected in species rank.
Principal component analysis of the vocal measurements underscores the vocal distinctness of Taliabu Grasshopper-Warbler L. portenta from the other three Wallacean species ( fig. S4 View Fig ). A cumulative proportion of variance of 73.3% is accounted for in the first two principal components, suggesting a strong vocal differentiation between Taliabu and the remaining taxa ( fig. S4 View Fig ).
Mitogenomic evidence: Using whole-genome sequencing and laboratory approaches for the use of ancient DNA, we created an alignment of whole-mitochondrial sequences spanning ~15,000 base pairs (bp) across 11 Locustella individuals belonging to all four constituent island forms of the L. castanea radiation, and used three different phylogenetic tree-building methodologies to investigate relationships and levels of divergence. More details on the methodology are provided below under separate Methods paragraphs.
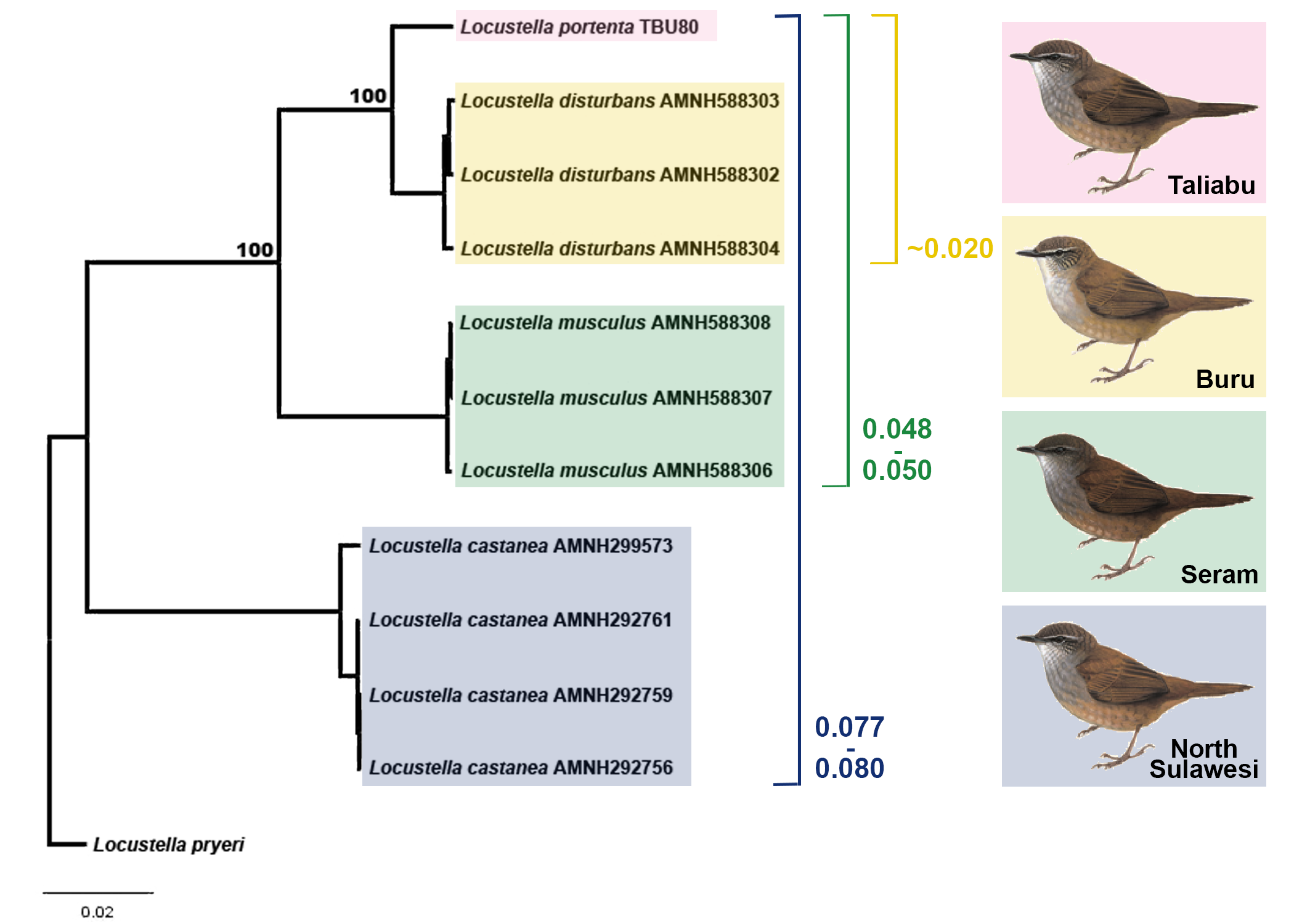
All three phylogenetic trees produced indicated identical topologies with high bootstrap support ( fig. S5 View Fig ), suggesting that the new L. portenta is an independent lineage most closely related to L. disturbans from Buru (mitogenomic divergence ~2%), and much more distantly related to L. musculus from Seram (divergence ~4.8–5%) and L. castanea from Sulawesi (divergence ~7.7–8%). Mitogenomic divergences roughly correlate with those from the COI barcoding gene, although they do not have to be equal in magnitude.
Integrating the vocal and mitogenomic evidence, it is clear that bioacoustic differences between L. portenta and L. disturbans , the two species which emerged most closely related on the mitogenomic tree ( fig. S5 View Fig ), far exceed those between L. portenta and a geographically distant species ( L. accentor ) that has never been closely associated with the L. castanea radiation. Our combined evidence therefore supports the treatment of L. portenta as a well-diverged small-insular endemic species.
Methodology
Bioacoustic sampling: We obtained 41 homologous recordings, which were either recorded by us in the field, or privately sourced from avid field ornithologists (including Robert O. Hutchinson and Ross Gallardy), or collected from publicly accessible online databases, namely the Avian Vocalization Center (AVoCet at http://avocet. integrativebiology.natsci.msu.edu), the Xeno-Canto Sound Library (www.xeno-canto.org), the Macaulay Sound Library (www.macaulaylibrary.org), and the Internet Bird Collection (www.hbw.com/ibc).
Although slight variation in recording quality exists in the equipment used, inspection and comparison of spectrograms from different devices showed that this variation is comparable to that amongst recordings made by one sole recordist using the same device. In addition, although there is some variability in background noise, spectrogram inspection suggested that measurements of vocalizations would not be affected. Measurements of the recordings were performed using the program Raven Pro Version 1.5 (Bioacoustics Research Program, Cornell Laboratory of Ornithology, Ithaca, NY, USA) on its default settings. The following eleven parameters were chosen for bioacoustic analysis, with each parameter averaged over all the measurements across one recording:
1. Total number of elements per motif (with an element being a syllable represented by a trace on the spectrogram, and a motif being a collection of elements always repeated together)
2. Number of introductory elements per motif (with introductory elements being brief, truncated, and usually fainter elements preceding the main body of the song)
3. Number of main elements per motif (with main elements taking up the bulk of the song, in Locustella typically in the form of shrill, insect-like trills)
4. Total duration of a motif
5. Total duration of introductory elements
6. Total duration of main elements
7. Duration of break between introductory and main elements
8. Minimum frequency of a motif
9. Maximum frequency of a motif
10. Bandwidth (i.e., maximum minus minimum frequency)
11. Peak frequency of a motif (i.e., the frequency at which the amplitude of the sound is
highest)
Bioacoustic analysis: The vocal diagnosability criterion outlined by Isler et al. ( 8 5), henceforth referred to as the Isler Criterion, was used to assess song diagnosability among taxa. Besides other oscine passerines ( 73, 8 6), this criterion has been employed as a bioacoustic species delimitation method across Asian birds such as owls ( 58, 87), pigeons ( 50, 52, 88), and nightjars ( 89). The Isler criterion has two parts which must be fulfilled in order to infer diagnosability. Firstly, the range of measurements of a parameter between two taxa must not exhibit overlap, and secondly, the data have to satisfy the following equation: xa + taSDa ≤xb–tbSDb, where ( x) refers to the respective means of measurements in taxon a and b, and ti refers to the t-score at the 95th percentile of the t distribution for n-1 degrees of freedom. This is a stricter statistical test than the t-test or Mann-Whitney U test ( 8 5) as it employs the standard deviation of the sample points instead of the taxon mean, allowing for the diagnosis of distinctions between taxa under more rigorous conditions. Successful Isler diagnosability is defined here as the presence of at least one vocally diagnostic parameter between taxa.
DNA extraction methods: Fresh tissue of the holotype of L. portenta was DNA- sequenced along with the following number of toepad scrapings of historical museum specimens: three of the Buru Grasshopper-Warbler L. disturbans , three of the Seram Grasshopper-Warbler L. musculus , and four of the Sulawesi Grasshopper-Warbler L. castanea . All museum samples had been kindly contributed by the American Museum of Natural History (for voucher numbers, see fig. S5 View Fig and table S5). DNA extraction was carried out with the Qiagen DNeasy Blood and Tissue Kit (Qiagen, Germany) following the manufacturer’s instructions. DNA extraction of the historic museum specimens’ toepad scrapings was conducted by trained personnel (CYG) in a room dedicated to ancient DNA extraction to preclude DNA contamination. The toepads were rinsed with water four times before extraction to ensure preservatives were removed, and DTT was added to facilitate digestion of keratin present in toepads. The concentrations of the DNA extracts were quantified using a high-sensitivity Qubit (Life Technologies, USA) assay.
DNA sequencing methods: The holotype’s DNA extract was sheared into smaller fragments of ~250 base pairs (bp) using 13 cycles of 30 seconds on and 30 seconds off on a Bioruptor Plus (Diagenode, USA). Whole-genome library preparation for the holotype was carried out using a NEBNext Ultra II DNA Library Prep Kit (New England Biolabs, USA) for Illumina according to the manufacturer’s protocol. For the museum toepad extracts, the shearing step was omitted as the ancient DNA is already degraded and NEBNext FFPE DNA repair mix (New England Biolabs, USA) was added to each ancient DNA sample prior to library preparation to fix damage common to degraded DNA such as deamination, nicks and gaps. In addition, size selection of library preparation products was omitted for the toepad samples to reduce DNA loss from bead selection. A total of five PCR cycles were run on the holotype’s DNA extract and 12 PCR cycles on the toepads’ extracts. Whole-genome library preparation of toepad samples was carried out in a room with a PCR hood dedicated to ancient library preparation work. The concentrations and peak fragment sizes of the libraries were quantified using a high-sensitivity Qubit (Life Technologies, USA) assay and an Advanced Analytical Fragment Analyzer (Agilent, USA), respectively. The peak fragment sizes of the historic museum samples and the fresh holotype (including adaptors and unique indexes) were ~230bp and ~380bp, respectively. All prepared libraries were sent to NovogeneAIT Genomics Singapore for Illumina sequencing using a HiSeq 4000 platform to produce 150bp paired-end reads.
Mitochondrial genome alignment: Trimmomatic (version 0.33) ( 90) was used to remove adaptors and filter low quality reads (below 15 Phred quality score). We applied the Burrows-Wheeler Aligner (version 0.7.15-r1140) ( 9 1) to align reads to a mitochondrial reference genome, Locustella pryeri (Genbank accession no.: NC_029151), with a minimum mapping quality score of 30. Samtools (version 1.9) ( 9 2, 9 3) was run to convert the SAM output files from the alignment to a BAM format for downstream analysis. Then Picard (version 2.18.21; http://broadinstitute.github.io/picard) was employed to insert read groups and remove PCR duplicate artifacts. The BAM files were then imported into CLC Genomics Workbench 7.0.4 (https://www.qiagenbioinformatics.com) for remapping to the same reference genome using the following mapping parameters: mismatch penalty of 1, insertion penalty of 1, and deletion penalty of 1. The reads were locally realigned at indel regions, and the consensus sequences were extracted at a minimum coverage threshold of 100. A mapping summary produced by Qualimap (version 2.2.1) ( 9 4) indicated that our mitochondrial genome coverage ranged between ~290–2920x for museum samples and ~6430x for the holotype (table S5). Sequences are deposited with GenBank at accession numbers MN597052 View Materials - MN597062 View Materials .
Phylogenetic analysis: The consensus sequences were exported in FASTA format and visually realigned in MEGA7 ( 95), producing a final whole-mitochondrial alignment of ~15,000bp. MEGA7 was used to construct a neighbor-joining tree based on uncorrected p- divergences and a maximum parsimony tree. Additionally, we ran raxmlGUI 1.5 beta ( 96) to construct a maximum likelihood tree using the GTRGAMMA model as specified by jModelTest (version 2.1.7) ( 97). All tree-building analyses were conducted with 10,000 bootstrap replicates. Pairwise uncorrected p-divergences across the mitogenomic alignment were calculated in MEGA7 with 10,000 bootstrap replicates.
No known copyright restrictions apply. See Agosti, D., Egloff, W., 2009. Taxonomic information exchange and copyright: the Plazi approach. BMC Research Notes 2009, 2:53 for further explanation.
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |