Abantiades, Herrich-Schaffer, 1855
|
publication ID |
https://doi.org/ 10.11646/zootaxa.4822.1.3 |
|
publication LSID |
lsid:zoobank.org:pub:AD57CF19-5F68-4992-943C-59ED442E2DD9 |
|
DOI |
https://doi.org/10.5281/zenodo.4452291 |
|
persistent identifier |
https://treatment.plazi.org/id/216587E0-0D2E-FF97-EFC4-F1EA3577FB83 |
|
treatment provided by |
Plazi |
|
scientific name |
Abantiades |
| status |
|
Systematics of Abantiades View in CoL
Molecular analyses
(see Fig. 1 View FIGURE 1 )
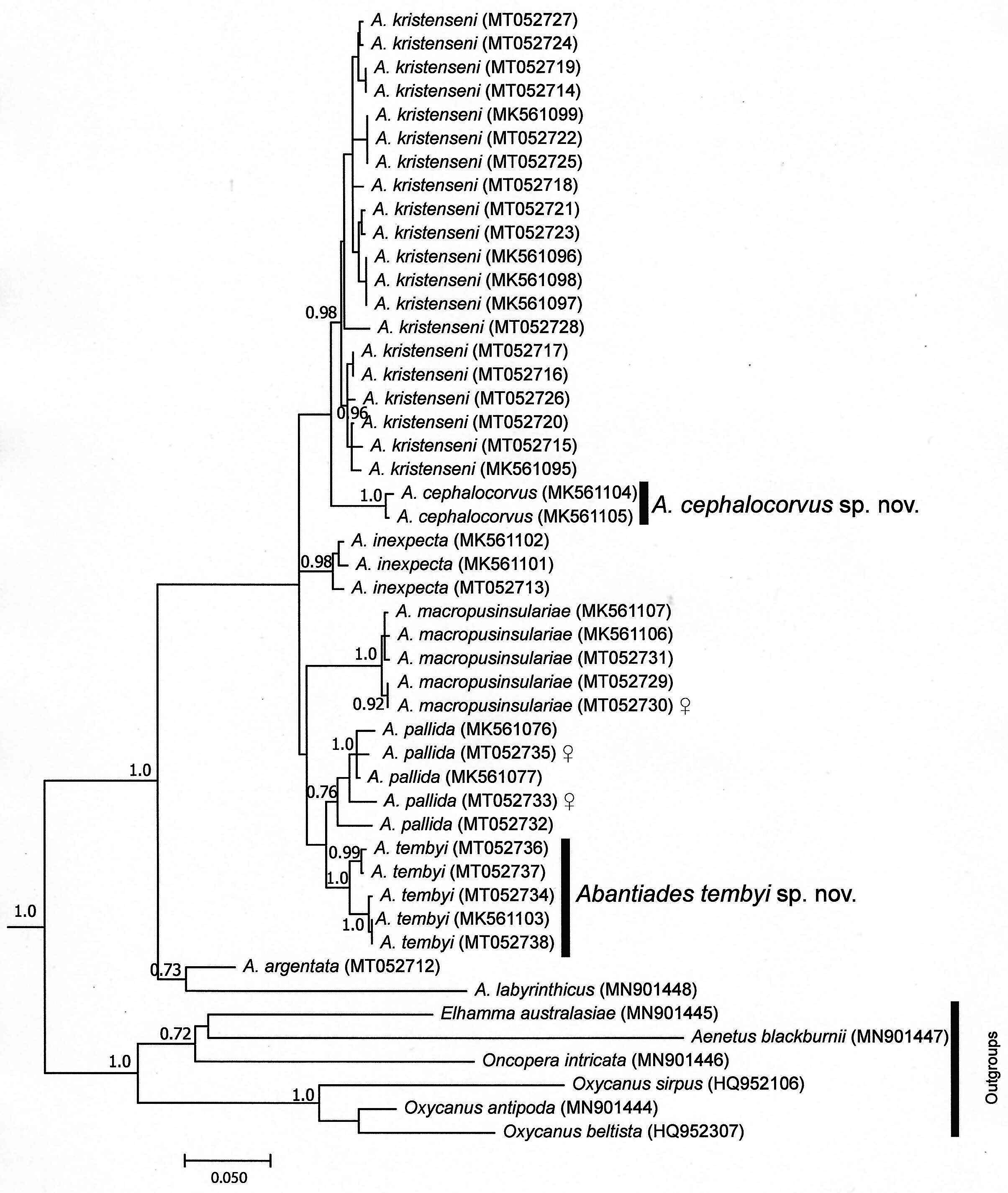
The Bayesian analysis for each species ( Fig. 1 View FIGURE 1 ) reveals a well supported (PP = 1.0) monophyletic short-beaked Abantiades clade compared to our six outgroup taxa. Although this is only based on the single mtDNA (COI) gene, it does corroborate the morphological distinction of the two new species from the six other Abantiades species in our analysis, which are each also defined by unique morphological characters (see Taxonomy section below). Supplementary Table S1 details all specimens used, including GenBank accessions and locality information. For instances where we obtained multiple individuals within species we found close intraspecific sequence divergences (Supplementary Table S2). For example, the greatest sequence divergence observed within any of our species varied between 0.6–3.3%, indicating variation that we would expect within species included here (Supplementary Table S2). These distances also correspond to those reported elsewhere for Lepidoptera that generally indicate below 3% intraspecific while above 3% interspecific comparisons using the same COI gene (e.g. Grund et al. 2019; Beaver et al. 2020). However, sequence divergences have been found to vary outside these ranges for some Abantiades species ( Simonsen et al. 2019; Moore et al. 2020).
Sequence divergence comparisons between all Abantiades species compared to our outgroup taxa were between 8.6–12.5% (Supplementary Table S2). Examining between Abantiades species reveal interspecific sequence divergences of up to 10.2% (between A. cephalocorvus sp. nov. and A. labyrinthicus (Donovan)) and up to 6.8% between the ‘triforked’ clade, which includes all Abantiades species in Fig. 1 View FIGURE 1 except A. labyrinthicus . We also observed several interspecific sequence divergences that are below 3%, for example A. pallida , A. kristenseni , and A. tembyi sp. nov. and A. inexpecta (Supplementary Table S2), which has been observed in other ghost moths (e.g. Moore et al. 2020). Although sequence divergence can be low between a number of Abantiades species, Fig. 1 View FIGURE 1 indicates that these are each monophyletic species (PP> 75%) that correspond to morphological differences (see Taxonomy section below). Comparisons between these trees and the sequence divergences of our new species described (see below) reveal that A. cephalocorvus sp. nov. is 3.9–5.0% divergent from its closest neighbouring species ( A. kristenseni ), while Abantiades tembyi sp. nov. differs from its closest neighbour ( A. pallida ) by 2.3–4.4%.
No known copyright restrictions apply. See Agosti, D., Egloff, W., 2009. Taxonomic information exchange and copyright: the Plazi approach. BMC Research Notes 2009, 2:53 for further explanation.
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |