Crocidura hikmiya, Meegaskumbura, Suyama, Meegaskumbura, Madhava, Pethiyagoda, Rohan, Manamendra-Arachchi, Kelum & Schneider, Christopher J., 2007
|
publication ID |
https://doi.org/10.5281/zenodo.180054 |
|
DOI |
https://doi.org/10.5281/zenodo.5622131 |
|
persistent identifier |
https://treatment.plazi.org/id/386A87B5-FFE7-266F-75C9-FE35FB7AFE37 |
|
treatment provided by |
Plazi |
|
scientific name |
Crocidura hikmiya |
| status |
sp. nov. |
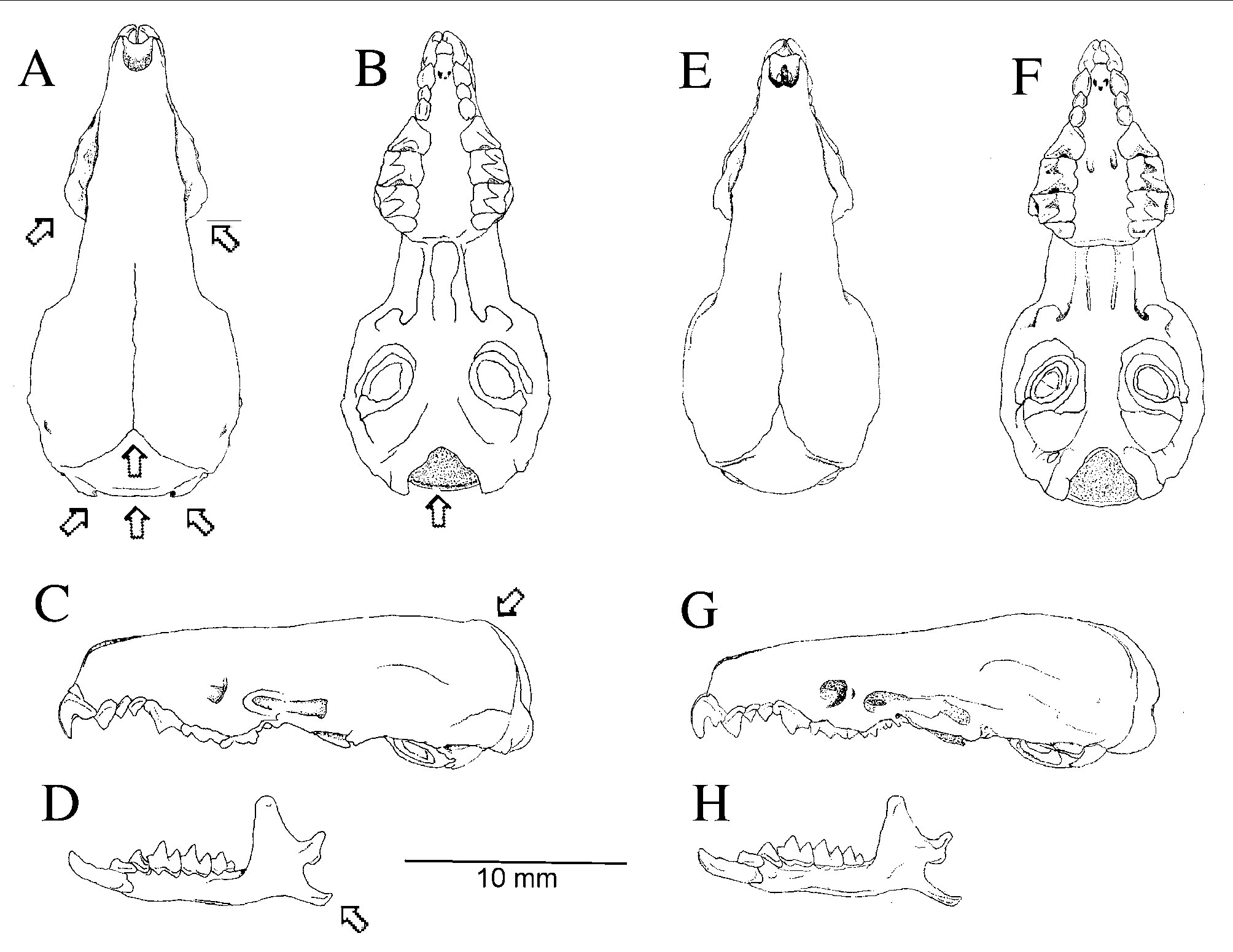
Crocidura hikmiya sp. nov. ( Fig. 2 View FIGURE 2 )
Holotype. WHT 6845 (male, 76.0 mm HBL, 7.8 g mass in life), preserved in alcohol; skull removed and preserved separately. Collected by M. Meegaskumbura and M. Bahir on 25 January 2004, at Kudawa, Sinharaja Forest Reserve ( 06°26’N, 80°25’E, elevation 460 m).
Paratypes. WHT 6836 (male, 64.0 mm HBL), WHT 6837 (female, 73.0 mm HBL), WHT 6843 (female, 69.9 mm HBL), and WHT 6844 (female, 71.0 mm HBL), collected from 20 – 25 January 2004, other collection data same as holotype. WHT 6849 (male, 62.5 mm HBL) and WHT 6853 (male, 67.1 mm HBL), collected by M. Meegaskumbura and K. Manamendra-Arachchi from 31 January- 4 February 2004 at Morningside ( 06°24.36’N, 80°86.87’E, elevation 1040 m). All specimens preserved in alcohol; skulls removed and preserved separately.
Etymology. The specific epithet ‘hikmiya’ is Sinhala for ‘shrew’, applied here as a substantive in apposition.
Diagnosis. Crocidura hikmiya is distinguished from C. miya , its sister-species (see Fig. 3 View FIGURE 3 and below), by the following external characters ( Table 1 View TABLE 1 ) and cranial characters ( Table 2 View TABLE 2 ): shorter tail length 101.4–114.5% of HBL ( vs 135.0–154.6% of HBL in C. miya ); longer forefeet, 9.3–10.2 mm ( vs. 8.7–9.0 mm in C. miya ); a narrower least inter-orbital breadth 3.9–4.2 mm ( vs. 4.2–4.4 mm in C. miya ); broader breadth of rostrum at narrowest point, 1.9–2.2 mm ( vs. 1.8–1.9 mm in C. miya ); occipital condyles protrude beyond braincase ( vs. condyles do not protrude beyond braincase in C. miya ); posterior edge of maxillary bone rounded ( vs. triangular in C. miya ); in dorsal view the occipital bone is triangular shaped with an obtuse angle ( vs. acute angle in C. miya ); occipital bone is slightly flattened on the back ( vs. rounded in C. miya ); in ventral view foramen magnum less deep ( vs. deep in C. miya ); dorsal posterior brain case is not smooth ( vs. smooth in C. miya ); angular process of dentary short and stout ( vs. long and thin in C. miya ) ( Fig. 4 View FIGURE 4 ). The two species also differ in coloration, C. hikmiya is dark grey-brown on the dorsum including the tail and slightly lighter colored on ventral surface of body and tail ( vs. C. miya which is brown on the dorsum including the tail and slightly lighter colored ventrally). There are no differences in the tooth characteristics between the two species.
Description. Crocidura hikmiya is a medium-sized shrew with a 64–76 mm HBL in the type series, which comprises sexually mature individuals. Tail slender, its length slightly exceeding that of head and body (tail length 101.4–114.5% of HBL) (see Table 1 View TABLE 1 ), semi-naked, with long, protruding hairs extending along proximal 13–24% of its length. Head length 26.2–28.9 mm (36.0–43.1% of HBL). Ears prominent, naked, height 7.0– 8.9 mm. Snout pink, with long bristles interspersed among shorter ones; bristles dark at base, paling to silvery grey at tips. Eyes small (< 1 mm in diameter, measured horizontally). General color of body including tail dark grey-brown. Venter slightly lighter colored than dorsum. Several long, dark, ‘guard hairs’ interspersed among grey hairs on both dorsal and ventral sides of body; individual hairs grey at base, with brown tips. Dorsal side of hands and feet semi-naked, appearing pinkish brown, with sparse brown hairs on digits.
External and cranial measurements (mm) of the holotype of C. hikmiya (WHT 6845): HBL, 76.0; HL, 27.8; TL, 77.5; HFL, 17.0; TBL, 19.7; FFL, 9.5; LAL, 12.4; EH, 8.9; GL, 20.2; BL, 18.2; BAL, 17.1; CL, 20.2; MTR, 6.0; PL, 8.8; PAL, 7.9; PPL, 9.8; LR, 7.1; BB, 9.0; LIOB, 4.2; PW1, 5.9; PW2, 3.1; BR1, 2.2; BR2, 6.1; BPM, 1.6; BMM, 1.0; HB, 5.2; ML, 11.4; LDI, 13.0; LDT1, 6.1; LDT2, 8.3; DD, 5.5.
Distribution. Crocidura hikmiya is known only from two forest-edge sites in Sinharaja Forest, at Kudawa and Morningside.
Variation. There is no apparent sexual dimorphism among the specimens collected. All specimens from both sites were of approximately the same size (mean mass = 7.7; SD = 1.0); however, both ventral and dorsal colors of the Morningside specimens were comparatively lighter than those from Kudawa. One specimen (WHT 6837) has 5 unicuspid teeth on each side of upper jaw instead of 3, as observed in all other specimens of C. hikmiya : the first of these is located immediately posterior to the first incisor, having thrusted the latter forwards and upwards. It has also displaced the tooth immediately posterior to it. The first incisor of this specimen is 0.2 mm smaller than the first incisor in other specimens. This ‘extra’ tooth is triangular in buccal and occlusal views and the pointed end protrudes towards the palate in occlusal view. The second ‘extra’ tooth is small, triangular in occlusal view and located at the end of the unicuspid tooth row filling the space between the other unicuspids and the pre-molar. Due to the addition of ‘extra’ unicuspid teeth, the length of unicuspid tooth-row has increased by 0.2–0.4 mm than other normal specimens. However, due to the smaller size of the first incisor and displaced unicuspid tooth posterior to the first ‘extra’ tooth, the upper jaw tooth row remains the same length as that of other specimens ( Fig. 5 View FIGURE 5 ).
Phylogenetic analysis. Both Bayesian and maximum parsimony phylogenetic trees based on mitochondrial DNA sequence data strongly supports C. hikmiya as the sister species of C. miya ( Fig. 3 View FIGURE 3 ). The Bayesian percentage posterior probability of the clade that includes only C. miya and C. hikmiya is 100, and the nonparametric parsimony bootstrap value for the same clade is 71% ( Fig. 3 View FIGURE 3 ). The percent pairwise uncorrected distance between these two species for the cytochrome- b gene fragment is 9.7–10.1%, while that between the currently-recognized species C. miya and C. horsfieldii is 11.5–11.6%; the latter, however, are not sister species. The percent pairwise uncorrected distance between C. hikmiya and C. horsfieldii is 13.3–13.6.
Morphological analysis. Principal Components Analysis of external measurements revealed that C. hikmiya and C. miya differ in multivariate morphological space along a single axis that represents tail length and forefoot length. PCA of the correlation matrix among variables resulted in four axes that explained a total of 88.7% of the total variance. Factor scores for C. hikmiya and C. miya overlap substantially on PC2, PC3, and PC4, but differ on PC1 ( Fig. 6 View FIGURE 6 A). PC1 explained 31.8% of the variance in external morphological variables. Tail length loaded heavily and positively on PC1, whereas forefoot length had a high negative loading. Factor scores from the two species do not overlap on PC1, with C. hikmiya having shorter tail and longer forefoot, and C. miya having longer tail and shorter forefoot.
PCA of cranial variables revealed that C. hikmiya and C. miya differed primarily on PC2, with substantial overlap of factor scores on PC1 ( Fig. 6 View FIGURE 6 B). PC1, which explained 33.6% of the variation, is largely a size axis, with high positive loadings for linear measurements reflecting skull dimensions (Basal Length, Mandible Length and Dentary Length Excluding Incisors). The two species overlapped substantially on PC1 indicating little variation in overall size. PC2 explained 20.1% of the total variance and had high positive loadings for Least Interorbital Breadth and Breadth of Braincase and high negative loading for breadth of rostrum at narrowest point and palatal length. Factor scores of the two species do not overlap on PC2, with C. hikmiya having a narrower least interorbital breadth, narrower breadth of braincase on average, broader breadth of rostrum at narrowest point and longer palatal length on average than C. miya ( Fig. 4 View FIGURE 4 ; Table 2 View TABLE 2 ). PC3 and PC4 explained 13.5% and 12.3% of the total variance respectively, but factor scores of the two species overlap completely on these axes (plots not shown). These analyses indicate that C. hikmiya and C. miya occupy different regions of morphospace for both cranial and external features and are diagnosable morphologically.
In addition, several characteristics of the skull also diagnose the species (see Diagnosis).
TABLE 2. Skull measurements of C. hikmiya and C. miya. Refer to Materials and Methods for abbreviations.
| C. hikmiya | C. miya | |||
|---|---|---|---|---|
| Range | Mean (SD) | Range | Mean (SD) | |
| GL | 19.2–20.2 | 19.7 (0.3) | 19.5–20.7 | 20.1 (0.5) |
| BL | 17.3–18.2 | 17.6 (0.3) | 17.3–18.0 | 17.6 (0.3) |
| BSL | 16.1–17.3 | 16.7 (0.4) | 16.4–17.2 | 16.8 (0.3) |
| CL | 19.2–20.2 | 19.7 (0.3) | 19.6–20.3 | 19.9 (0.3) |
| MTR | 5.6–6.1 | 5.8 (0.2) | 5.6–5.8 | 5.7 (0.1) |
| PL | 8.3–8.8 | 8.6 (0.2) | 7.9–8.4 | 8.1 (0.2) |
| PAL | 7.2–7.9 | 7.5 (0.2) | 7.1–7.6 | 7.4 (0.2) |
| PPL | 9.2–9.8 | 9.5 (0.2) | 9.2–10.1 | 9.7 (0.4) |
| LR | 6.9–7.1 | 7.0 (0.1) | 6.8–7.3 | 7.0 (0.2) |
| BB | 8.9–9.0 | 9.0 (0.1) | 8.9–9.2 | 9.1 (0.1) |
| LIOB | 3.9–4.2 | 4.1 (0.1) | 4.2–4.4 | 4.3 (0.1) |
| PW1 | 5.5–5.9 | 5.7 (0.2) | 5.7–5.9 | 5.8 (0.1) |
| PW2 | 2.6–3.1 | 2.8 (0.2) | 2.7–2.9 | 2.8 (0.1) |
| BR1 | 1.9–2.2 | 2.0 (0.1) | 1.8–1.9 | 1.9 (0.1) |
| BR2 | 5.9–6.2 | 6.1 (0.1) | 6.0–6.2 | 6.1 (0.1) |
| BPM | 1.4–1.6 | 1.5 (0.1) | 1.4–1.6 | 1.6 (0.1) |
| BMM | 1.0–1.1 | 1.0 (0.0) | 0.9–1.1 | 1.0 (0.1) |
| HB | 4.8–5.2 | 4.9 (0.1) | 4.9–5.2 | 5.1 (0.1) |
| ML | 10.8–11.4 | 11.1 (0.2) | 10.5–11.0 | 10.8 (0.2) |
| LDI | 12.4–13.1 | 12.8 (0.2) | 12.2–12.9 | 12.5 (0.3) |
| LDT1 | 6.0–6.2 | 6.1 (0.1) | 6.0–6.3 | 6.1 (0.1) |
| LDT2 | 8.0–8.3 | 8.1 (0.2) | 7.9–8.3 | 8.1 (0.2) |
| DD | 5.2–5.6 | 5.4 (0.1) | 5.0–5.4 | 5.2 (0.2) |
No known copyright restrictions apply. See Agosti, D., Egloff, W., 2009. Taxonomic information exchange and copyright: the Plazi approach. BMC Research Notes 2009, 2:53 for further explanation.