Anoectochilus roxburghii subsp. glycosyltransferases, (Wall.) Lindl. (Wall.) Lindl.
|
publication ID |
https://doi.org/ 10.1016/j.phytochem.2021.113007 |
|
DOI |
https://doi.org/10.5281/zenodo.8239952 |
|
persistent identifier |
https://treatment.plazi.org/id/70107B13-F650-8567-FFDD-50D2401EFB9B |
|
treatment provided by |
Felipe |
|
scientific name |
Anoectochilus roxburghii subsp. glycosyltransferases |
| status |
|
2.2. Screening of A. roxburghii glycosyltransferases in the TX-TL system
After showing the capability of this approach in screening UDPglucose glycosyltransferases of the model plant A. thaliana , we applied the same approach on a local medicinal plant A. roxburghii in Fujian, China, to screen its glycosyltransferases which play an essential role in the synthesis of its biologically active natural products. To obtain the sequence information of all the putative UDP-glucose glycosyltransferases, we performed transcriptome sequencing on this plant. The transcriptome data of A. roxburghii were obtained by Illumina HiSeqTM 2000 sequencing in our previous study ( Zou et al., 2019). First, we extracted the total RNA of A. roxburghii , and mRNA was enriched by Oligo (dT) beads. Then the enriched mRNA was fragmented into short fragments using fragmentation buffer and reverse transcribed into cDNA with random primers. Second-strand cDNA was synthesized by DNA polymerase I, RNase H, dNTP, and buffer. Then the cDNA fragments were purified with QiaQuick PCR extraction kit, end repaired, poly(A) added, and ligated to Illumina sequencing adapters. The ligation products were size-selected by agarose gel electrophoresis, PCR amplified, and sequenced using Illumina HiSeqTM 2000. Reads containing adapters, reads containing more than 10% of unknown nucleotides (N), and low-quality reads were removed from the raw reads. The resulting clean reads were further assembled to identify high-quality transcripts. Transcriptome de novo assembly was carried out with a short-read assembling software Trinity. A total of 58,065 transcripts were assembled with a mean length of 806 bp and an N50 of 1347 bp. Thus, these data provide a good foundation for the subsequent screening of putative UDP-glucose glycosyltransferases ( Zou et al., 2019). Then, we used the BLASTx program with an E-value threshold of 1e 5 to the Kyoto Encyclopedia of Genes and Genomes (KEGG) database, and protein functional annotations could be obtained. Since each sequence alignment result may have more than one hit, in order to ensure its biological significance, an optimal alignment result was retained as the annotation of the gene. A total of 96 UDP-glucose glycosyltransferase related genes were discovered using the KEGG database. Finally, we used the getORF software to filter 27 glycosyltransferases with an open reading frame (ORF) ( Wang et al., 2019; Zhang et al., 2021) ( Table 2 View Table 2 ).
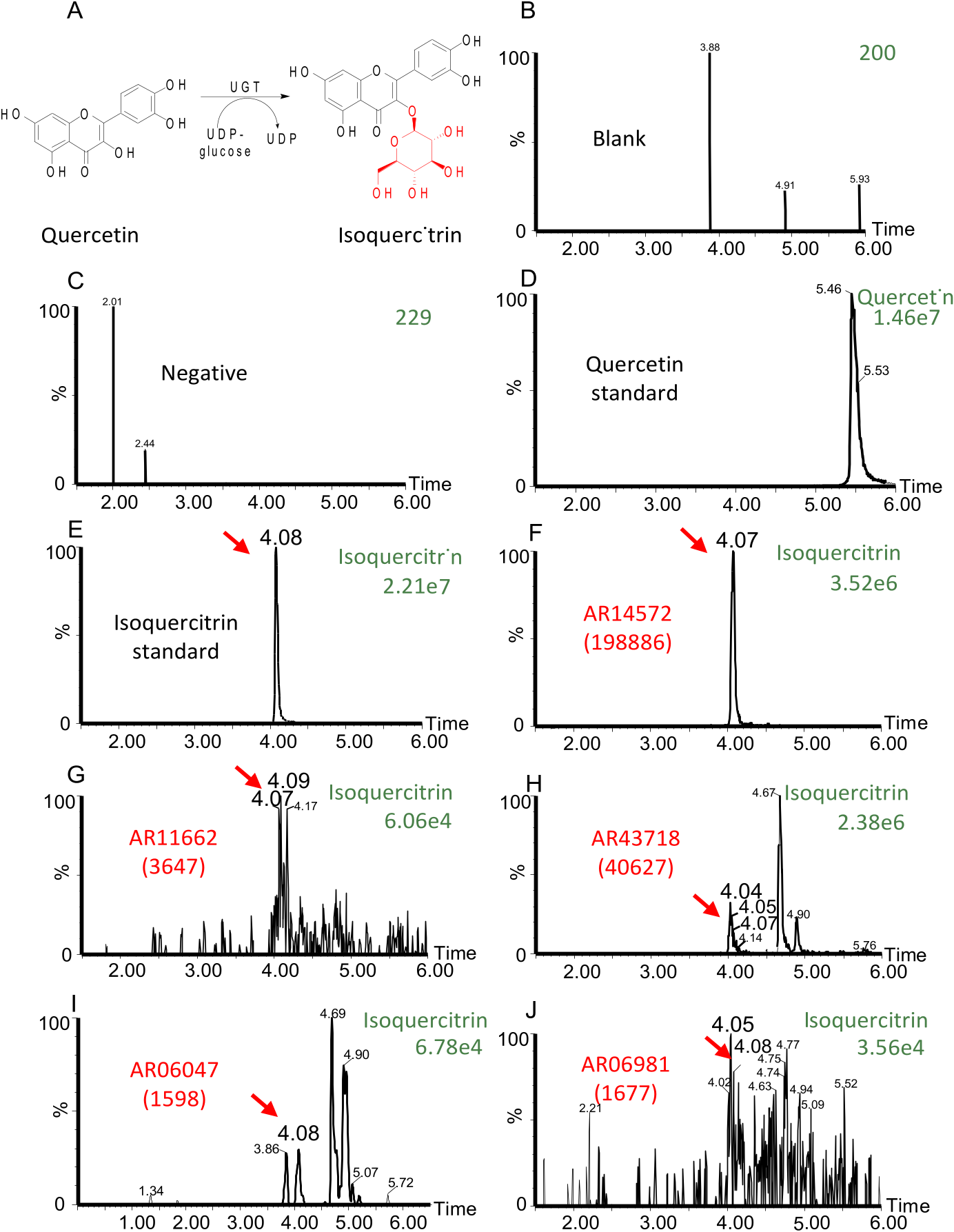
We then used the reverse-transcribed cDNA from A. roxburghii as the template and amplified these ArUGTs genes using PCR. We successfully generated 11 different linear DNAs ready for the TX-TL reaction. Then we expressed the corresponding glycosyltransferases in TX-TL and tested their glycosylation activity with quercetin as it is the most representative flavonoid aglycone. After analyzing reaction products using UPLC-MS, we found that 6 ArUGTs possessed significant glycosyltransferase activity in TX-TL reaction solutions (See Fig. 3 View Fig , Supplementary Fig. S5 View Fig ).
Comparison of the chromatogram of quercetin standard, the chromatogram of isoquercitrin standard, and the chromatograms of the glycosylated products showed that these 6 ArUGTs – AR06047, AR06981, AR07558, AR11662, AR14572, and AR43718 – can catalyze the conversion of quercetin to isoquercitrin ( Fig. 3 View Fig ). In contrast, the other 5 ArUGTs (AR07805, AR40806, AR47165, AR14280, AR42458) did not show any significant glycosyltransferase activity. As enzymes have substrate specificity, the main purpose of this study is to establish and verify a method for rapid screening of plant UGTs. A. roxburghii is known to contain a variety of quercetin glycosides, so for the time being we only chose quercetin as a substrate to screen for new enzyme genes. Those putative enzymes that cannot catalyze quercetin may have catalytic activity on other substrates. We will further expand the scope of substrate screening in follow-up studies. As the positive results, we have successfully screened out 6 new ArUGTs which showed significant glycosyltransferase activity to substrate quercetin with this potential strategy among 11 different linear DNAs expressed in the TX-TL reaction (Supplementary Table S2 View Table 2 ).
No known copyright restrictions apply. See Agosti, D., Egloff, W., 2009. Taxonomic information exchange and copyright: the Plazi approach. BMC Research Notes 2009, 2:53 for further explanation.
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |