Eunapius fragilis ( Leidy, 1851 )
|
publication ID |
https://doi.org/10.5852/ejt.2022.828.1861 |
|
publication LSID |
lsid:zoobank.org:pub:92E8908C-818C-49CD-8E84-A95479F8E455 |
|
DOI |
https://doi.org/10.5281/zenodo.6841099 |
|
persistent identifier |
https://treatment.plazi.org/id/90618787-FFC4-FFFD-FDC1-FDCC1C9391FE |
|
treatment provided by |
Felipe |
|
scientific name |
Eunapius fragilis ( Leidy, 1851 ) |
| status |
|
Eunapius fragilis ( Leidy, 1851) View in CoL
Fig. 5 View Fig
Spongilla fragilis Leidy, 1851: 278 View in CoL .
Material examined ( 11 specimens, Table 1 View Table 1 )
SWEDEN – Uppland • 1 spec.; Hammarskogs bath ; 59.7636° N, 17.5762° E; 2 Aug. 2017; Chloé Robert and Raquel Pereira leg.; UPSZMC 188108 GoogleMaps • 2 specs; Flottsund ; 59.7875° N, 17.6626° E; 2 Aug. 2017; Chloé Robert and Raquel Pereira leg.; UPSZMC 188116 , 188118 . – GoogleMaps Jämtland • 8 specs; Lake Storsjön, Galhammar ; 62.7784° N, 14.4321° E; 30 Aug. 2017; Chloé Robert and Raquel Pereira leg.; UPSZMC 188163 , 188215 , 188217 , 188219 , 188221 , 188223 , 188225 , 188227 GoogleMaps .
Description
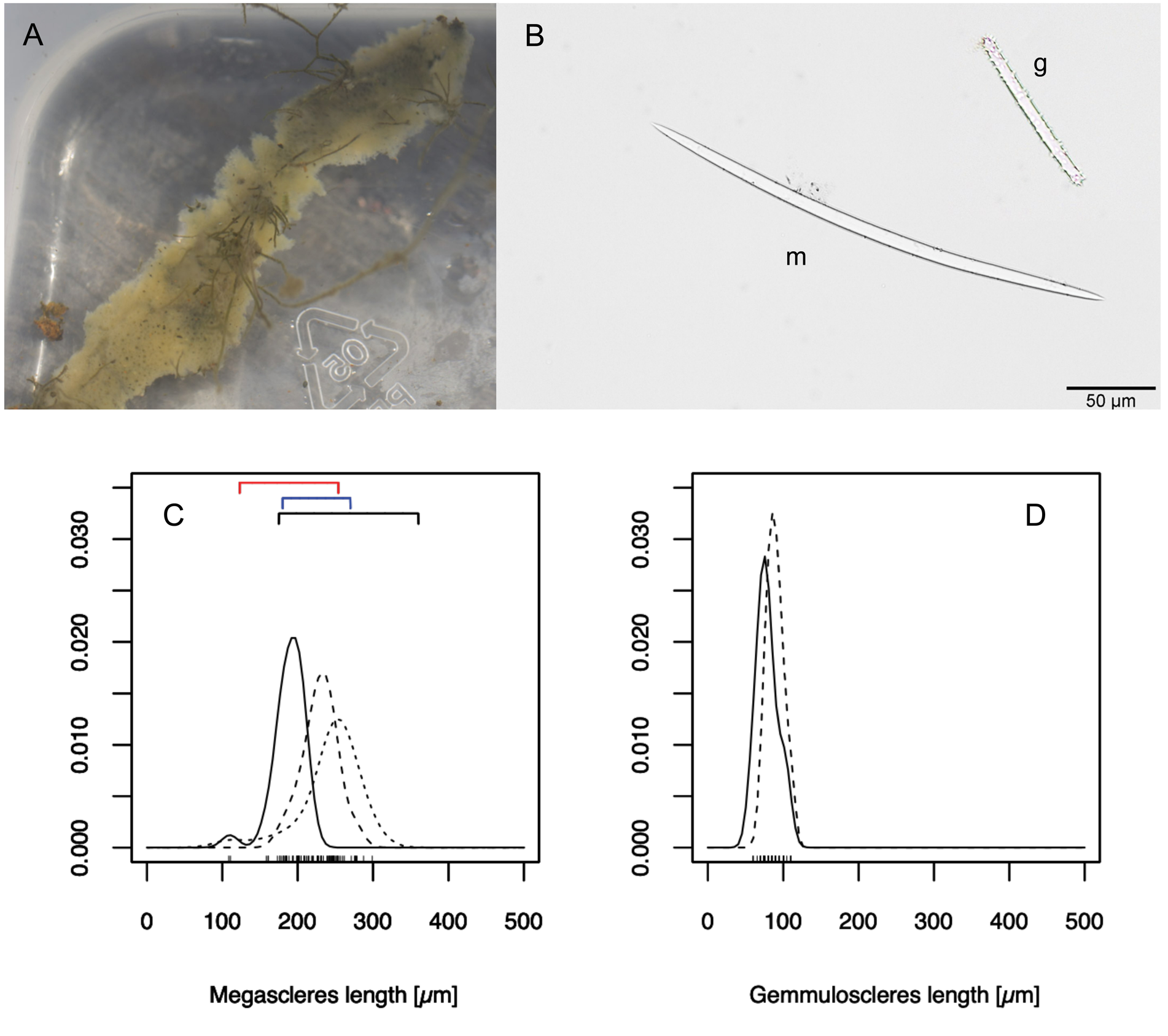
HABITUS. Specimens of this species have a pale green colour, slightly lighter than the other three species ( Fig. 5A View Fig ).
SPICULAE. Megascleres and gemmuloscleres. Megascleres are smooth and fusiform oxeas ( Fig. 5B View Fig ). Gemmuloscleres are spiny and rod-shaped ( Fig. 5B View Fig ). Measured megascleres 219.5 μm (110 to 300 μm) long ( Fig. 5C View Fig ) and 6.7 µm (2.5 to 10 μm) wide. Few gemmuloscleres were present, 49 in two specimens were measured. Their average length was 85.5 μm (60 to 110 μm) ( Fig. 5D View Fig ) and width 5.6 μm, (2.5 to 7.5 μm).
Distribution and habitats
Specimens collected in southeast and northern part of sampling area ( Fig. 1 View Fig ); no specimens found in the western part; from lakes and in a river close to lake inlet.
Bar-code analyses
There was a high within-species similarity in the specimens of the present study, and they fall well within the variation encountered in published sequences. Particularly cox I sequences were useful ( Fig. 6A View Fig ), whereas the use of 28S ( Fig. 6B View Fig ) was hampered by fewer available sequences in the databases and that the available sequences in many cases were short; of the ca 660 bp amplified by the used primers, there was only ca 350 bp of useful overlap with most GenBank sequences. Both markers provided useful data to identify the species.
Marker search
Primer pairs tested for amplification are listed in Supp. file 1, but most of them failed to amplify S. lacustris DNA extracts. Different annealing temperatures and MgCl 2 concentrations were used, with the latter having no effect on the amplification results. No product was detected for 21 of the 24 primer pairs tested. The remaining three primer pairs were successful; i56-F/i56-R, i56-Spla-F/i56-Spla-R and ATP6porF/ATP6porR (primer pairs 23, 25 and 5 in Supp. file 1). The marker i56 is an intron located in a conserved gene coding for glutamyl-prolyl-tRNA-synthetase ( Chenuil et al. 2010). Using the primer pair i56-F/i56-R at an annealing temperature of 64°C (following tests of optimal temperature) product was obtained for 43 samples, and 40 of these were successfully sequenced.
The primer pair ATP6porF/ATP6porR, targeting the mitochondrial gene ATP6, amplified a region ca 450 bp long. However, as expected, a low amount of variation was detected between specimens, with less than 0.7% nucleotide variation. Thus, for this study, only primer pair i56-F / i56-R was selected.
Genetic diversity
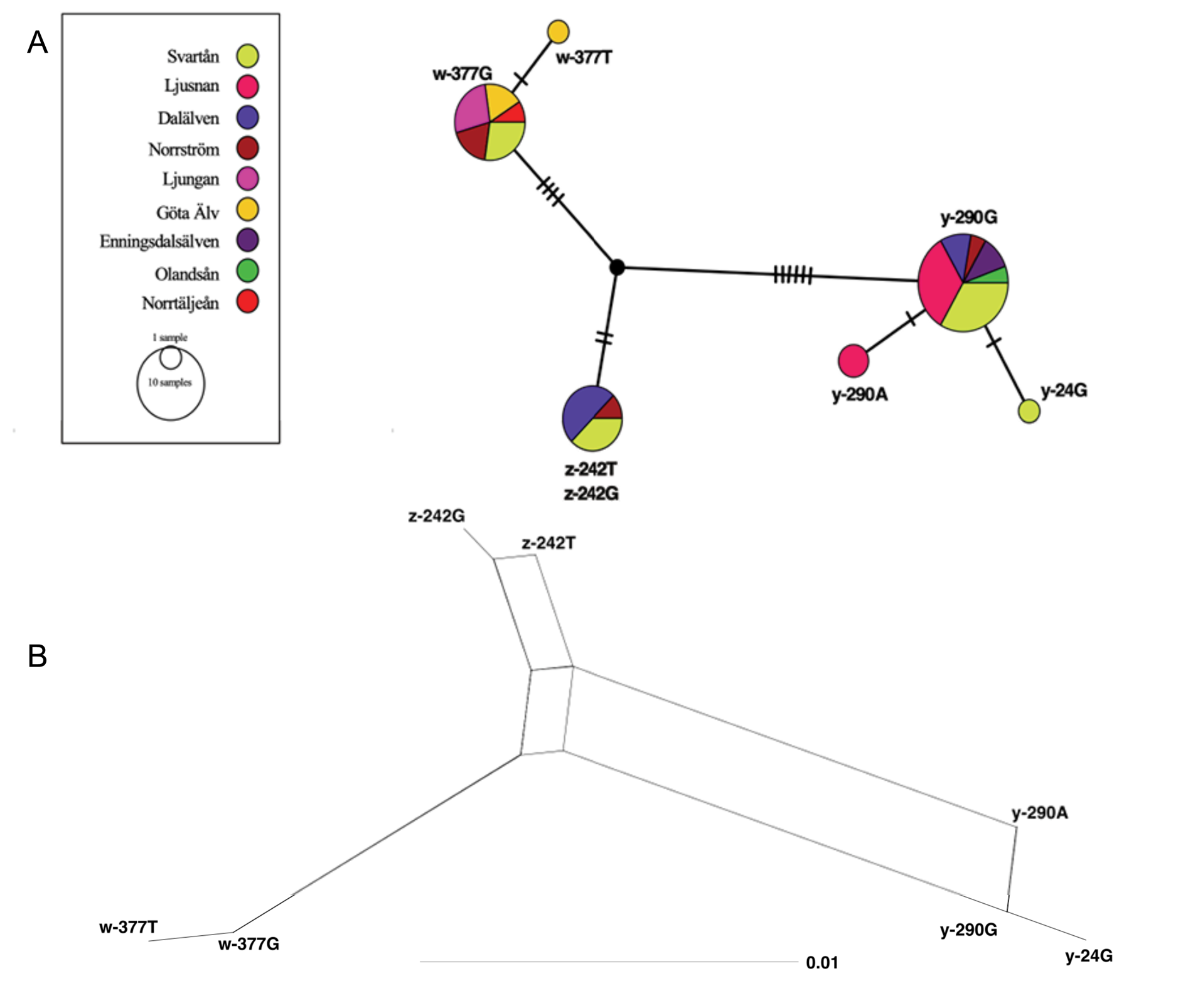
For the i56 marker we found seven distinct sequence variants in the sampled S. lacustris populations, which we designate y-24G, y-290G, y-290A, z- 242T, z-242G, w-377G and w-377T (see Table 3 View Table 3 ). The most abundant variant was y-290G, observed for 18 individuals collected in six different catchment areas. Variants w-377G and z-242T were also common; 11 and 7 specimens, respectively, shared these variants, sampled from five and three different places. Remaining variants (y-24G, y-290A, w-377T, z-242G) were found only for one or two specimens. Two specimens from the Ljusnan catchment area shared variant y-290A, one individual from Dalälven had variant z-242G, one from Göta Älv had w-377T, and a single individual from the Svärtaån catchment area was found with y-24G. Genetic distance and raw differences between variants are shown in Table 4 View Table 4 . Variants y-290A and w-377T are least similar, with 24 segregating sites and a 2.7% HKY85 nucleotide difference. Least difference distance is found between y-290G and y-290A, z-242T and z-242G, and w-377G and w-377T with just one segregating site.
Phylogenetic network
In a phylogenetic network we see three main clusters; y-24G and y-290G with y-290A, w-377G with w-377T, and z-242T with z-242G ( Fig. 7 View Fig ).
As PopArt does not handle gapped sites, parts of the alignment containing gaps were thus ignored in the analysis. Thus, in the median joining network, variants z-242T and z-242G are seen as identical ( Fig. 7A View Fig ) as they differ only in indel sites. In the majority of catchment areas (Svärtaån, Norrström, Göta
Älv, Ljusnan and Dalälven) several variants are present. Only one variant is found in Enningdalsälven and Ljungan, where two and three animals, respectively, were assessed.
Variants y-290G and w-377G are distributed over the entire study area, whereas variant z-242T is restricted to the middle-west catchment areas (see Fig. 8 View Fig ). Interestingly, only these three variants (y-290G, w-377G and z-242T) were found in the west part of Sweden. Variants y-290A, w-377T and z-242G are found in the same catchment areas as their closest allelic counterpart (y-290G, w-377G and z-242T, respectively).
Results of AMOVA (analysis of molecular variance) show more variation within than among populations (59.8% and 40.2% of the variation, respectively).
No known copyright restrictions apply. See Agosti, D., Egloff, W., 2009. Taxonomic information exchange and copyright: the Plazi approach. BMC Research Notes 2009, 2:53 for further explanation.
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
SubClass |
Heteroscleromorpha |
|
Order |
|
|
Family |
|
|
Genus |
Eunapius fragilis ( Leidy, 1851 )
| Robert, Chloé, Pereira, Raquel & Thollesson, Mikael 2022 |
Spongilla fragilis
| Leidy J. 1851: 278 |