Hieracium, L.
|
publication ID |
https://doi.org/ 10.1016/j.phytochem.2018.06.008 |
|
DOI |
https://doi.org/10.5281/zenodo.10513942 |
|
persistent identifier |
https://treatment.plazi.org/id/90640E19-FFDE-8D46-FCF0-8012FBDB47D5 |
|
treatment provided by |
Felipe |
|
scientific name |
Hieracium |
| status |
|
2.2. Qualitative and quantitative LC-MS analyses of SLs in the dry methanol extracts of flowering aerial parts of investigated Hieracium View in CoL species and statistical analysis of obtained data
The LC-MS analysis of the dry methanol extracts of flowering aerial parts of 28 Hieracium species was performed using a method optimized for the separation of water-soluble amino acid-SL conjugates. The identification of SLs in analysed extracts was done by the comparison of their retention times and MS spectra to those of isolated SLs (1–4), used as reference compounds. Their quantification in the extracts was performed by the external standard method, using the peak areas of the SIM chromatograms obtained by the monitoring of [M+H] + at m/z 540.20 (1 and 3) and at m/z 378.10 (2), and [M+NH 4] + at m/z 442.20 (4) ( Figs S5.2 and 5.4 View Fig ). The regression equations of the calibration curves, their correlation coefficients (r 2), concentration ranges, limits of detection (LODs) and quantification (LOQs) are given in Table S2 View Table 2 . The quantities of identified SLs in analysed dry extracts, expressed as mg/g of extract, are presented in Table 3 View Table 3 .
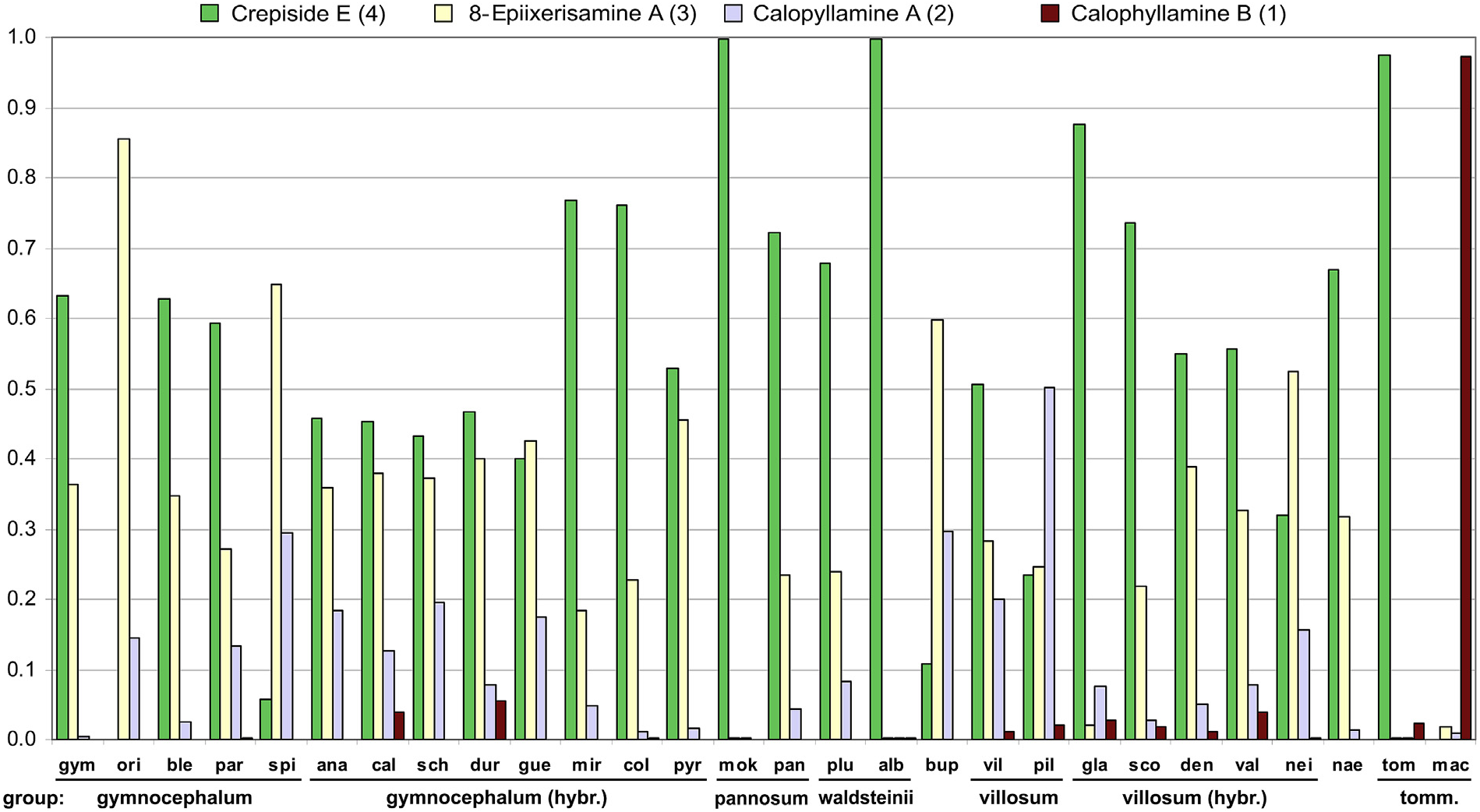
Crepiside E (4) was detected in 26 species, in 21 of them it was the most abundant SL, and it was not detected only in H. orieni A. Kern. and H. macrodontoides (Zahn) Zahn. Its quantities broadly ranged from 0.26 mg /g in H. pilosum Froel. extract to 15.57 mg /g in H. blecicii Niketi ć extract. 8-Epiixerisamine A (3) and calophyllamine A (2) were present in all of the investigated species (the first was dominant in 5, and the second in one species). Their content varied from traces in the extracts of the same five species, to 11.14 mg /g in H. guentheri-beckii Zahn extract (3), and to 5.31 mg /g in H. anastrum (Degen & Zahn) Niketi ć extract (2) ( Table 3 View Table 3 ).
The only identified eudesmanolide-amino acid conjugate, calophyllamine B (1), was detected in only 14 species, where it was present in low quantities, ranging from traces in two extracts to 0.88 mg /g in H. calophyllum extract. Calophyllamine B was dominant SL only in H. macrodontoides extract ( Table 3 View Table 3 ).
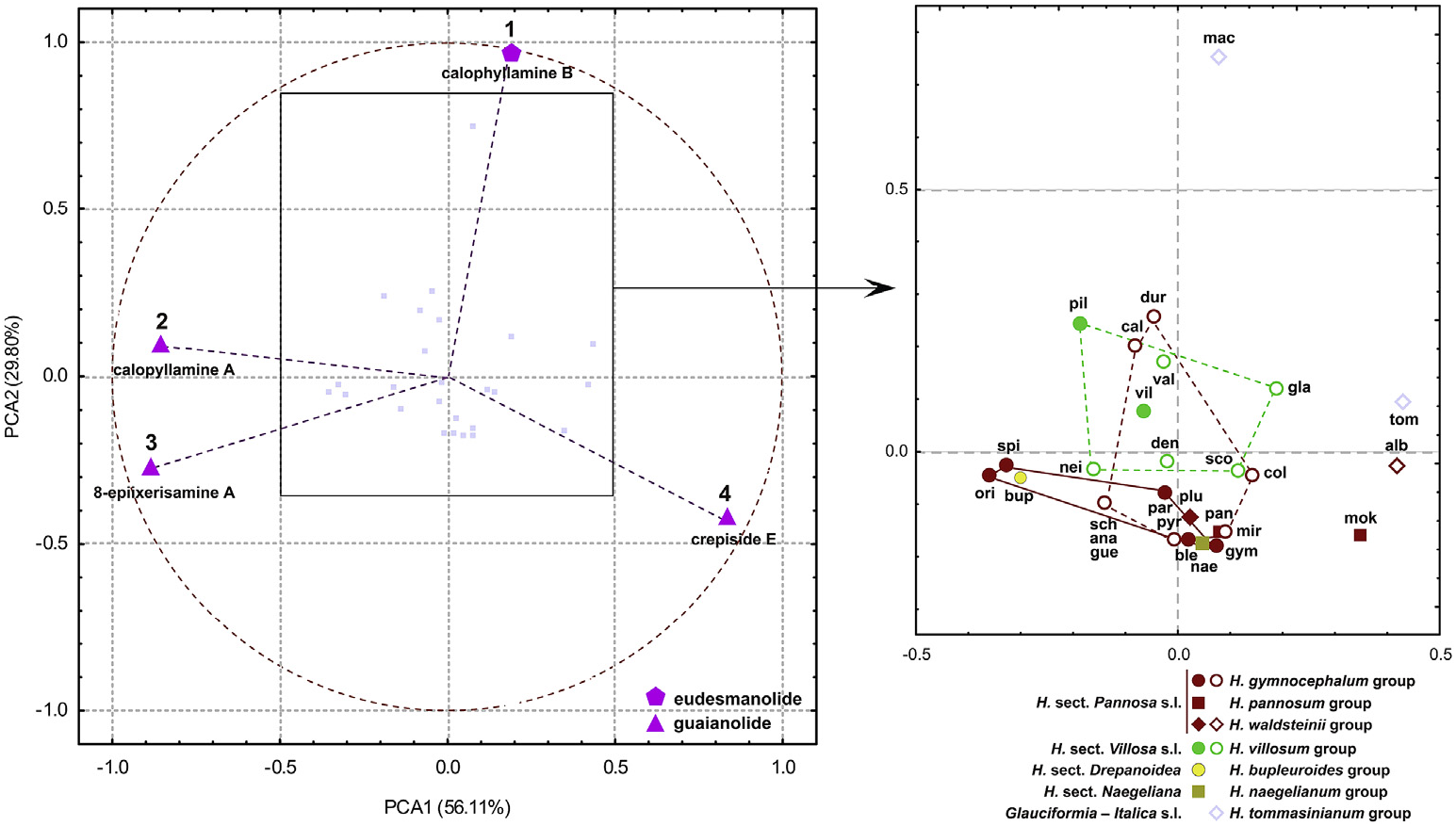
In order to compare the analysed species, the percentages of SLs relative to total detected SLs in each extract were presented ( Fig. 3 View Fig ). To find compounds that significantly contribute to differentiation between species, principal component analysis (PCA) was performed. Results showed that all four SLs were highly correlated with variance, regardless of their average contents. Three guaianolides had the highest loadings along the first principal axis which explained 56.32% of the total variance ( Fig. 4 View Fig ). Crepiside E (4) was positively (0.83), while 8- epiixerisamine A (3) (−0.89) and calopyllamine A (2) (−0.86) were negatively correlated. Eudesmanolide calophyllamine B (1) mostly contributed to the second axis, which explained smaller amounts of the data variation (29.80%). However, this compound was remarkably negatively correlated with the factor 2 (−0.96) rather contributing to it (77.98%). Therefore, all four SLs were more or less equally important for the variation, and they were distributed near the correlation circle, not deviating too far from orthogonality.
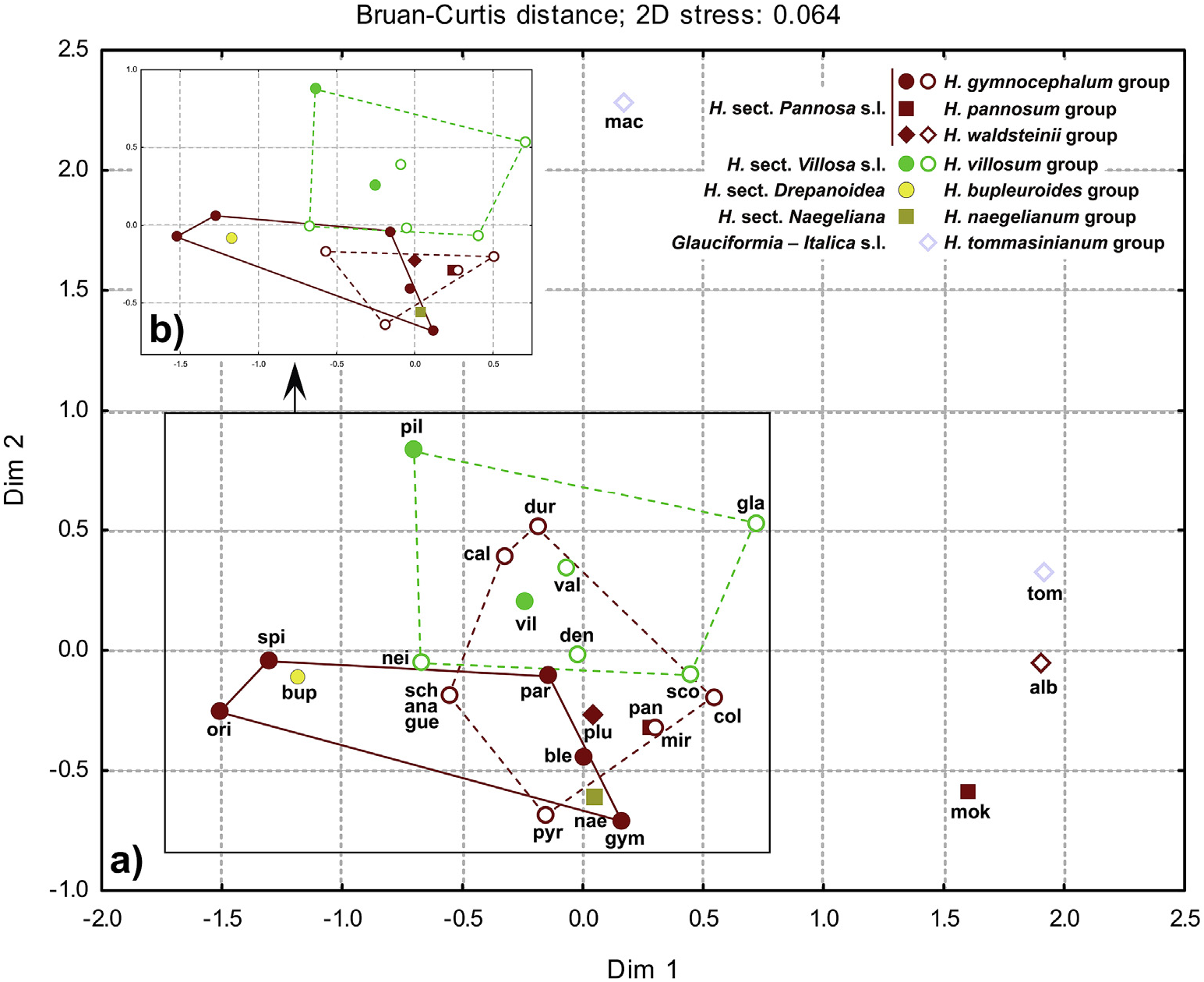
The majority of the species were clustered near the coordinate origin in PCA, as well as in nMDS, with relative SLs content decreasing in the following order: crepiside E (4), 8-epiixerisamine A (3), calophyllamine A (2), and calophyllamine B (1) ( Figs. 4–5 View Fig View Fig ). The species that did not fully follow this order, containing much more 8- epiixerisamine A (3) than crepiside E (4) ( H. orieni , H. spirocaule Niketi ć and H. bupleuroides C. C. Gmel. s.l.) or an extremely large relative amount of crepiside E (4) ( H. mokragorae (Nägeli & Peter) Nägeli & Peter , H. albopellitum (Zahn) Niketi ć and H. tommasinianum K. Malý ) ( Fig. 3 View Fig ), were located on the opposite sides of the first axis ( Figs. 4–5 View Fig View Fig ). Hieracium pilosum was the only species with the highest relative content of calophyllamine A (2) ( Fig. 3 View Fig ). Guaianolides were the dominant SLs in almost all of the investigated species. Only H. macrodontoides , clearly separated along the second axis ( Figs. 4–5 View Fig View Fig ), contained a remarkably high percentage of eudesmanolide calophyllamine B (1) ( Fig. 3 View Fig ), although in a low absolute content ( Table 3 View Table 3 ). Despite being detected in low concentrations in all of the investigated species, this SL could be a good chemosystematic marker for some groups, due to its absence in the basic sections Pannosa (Zahn) Zahn , Drepanoidea Monnier and Naegeliana Zahn ex Szel ą g, as well as in more than a half of hybridogenous species that originated from H. sect. Pannosa. On the contrary, this SL was almost always present in H. sect. Villosa (Griseb.) Gremli (incl. hybridogenous species, except H. neilreichii Beck ) and Glauciformia (Freyn) Zahn – Italica (Fr.) Av. Touv. ( H. tommasinianum group).
PCA and nMDS scores of the investigated species and the main taxonomic groups in many cases reflected differences between taxa ( Figs. 4 View Fig and 5a View Fig ). Their general positions were similar to those from the PCA and nMDS analyses of flavonoids and phenolic acids in investigated Hieracium species (Milutinovi ć et al., 2018), but with evident overlaps between groups. After eliminating the scores of two hybridogenous species ( H. calophyllum and H. durmitoricum (Rohlena & Zahn) Niketi ć) in nMDS, the overlap was not particularly significant and the representatives of sections Pannosa and Villosa (incl. hybridogenous species) were well separated along the second axis ( Fig. 5b View Fig ). These differences could be explained by the lack of eudesmanolide calophyllamine B (1) in section Pannosa (except in traces in H. paratrichum Niketi ć and H. albopellitum ) ( Fig. 3 View Fig ). This compound was also not detected in H. bupleuroides group (sect. Drepanoidea ) and H. naegelianum Pan č i ć (sect. Naegeliana) ( Fig. 3 View Fig ), and consequently these species had a similar position as the representatives of H. gymnocephalum Griseb. ex Pant. group s.s. ( Figs. 4–5 View Fig View Fig ).
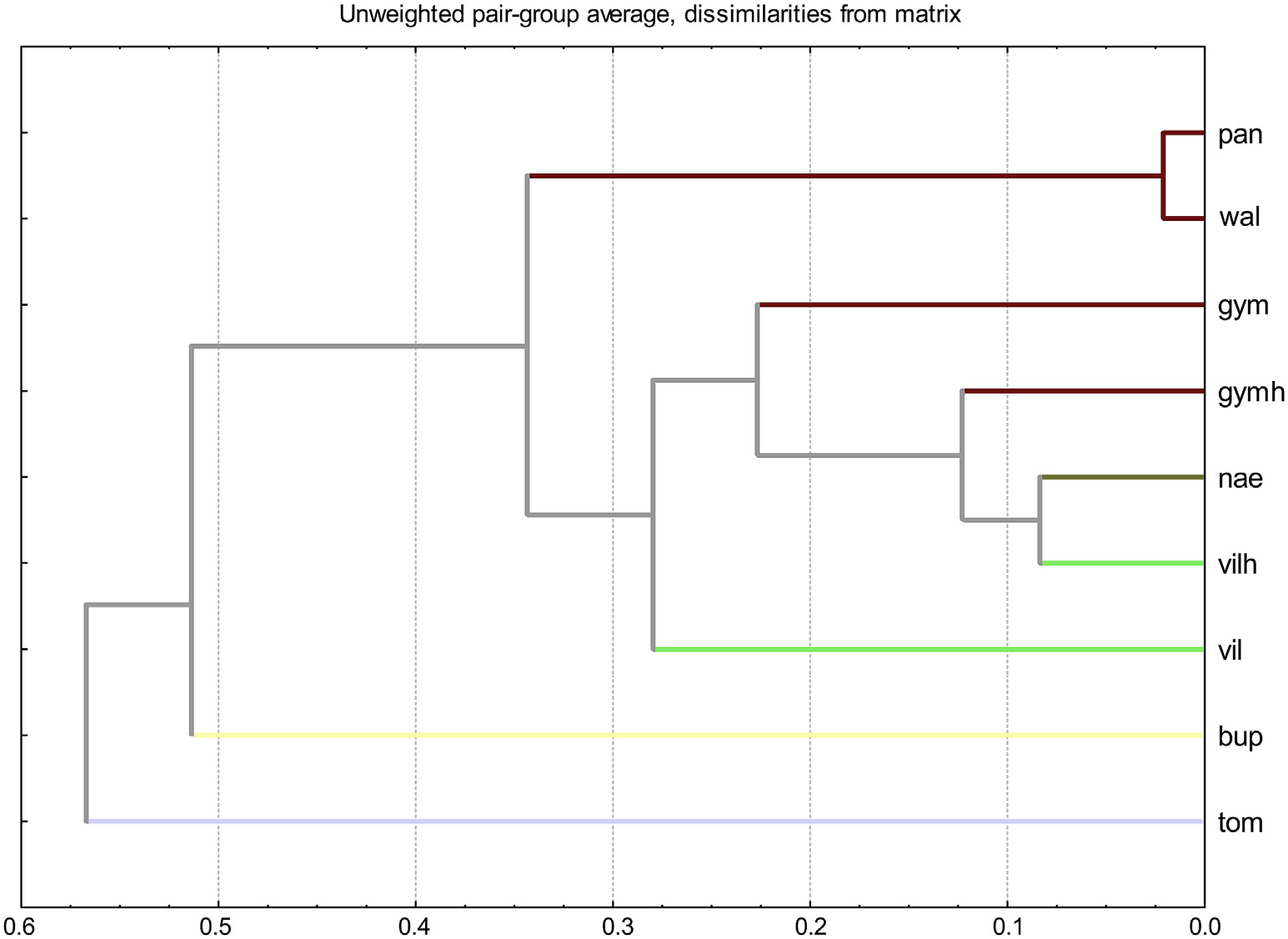
UPGMA dendrogram of the investigated groups ( Fig. 6 View Fig ) showed that Glauciformia–Italica ( H. tommasinianum group), particularly influenced by the isolated position of H. macrodontoides , was clearly separated from other sections. Section Drepanoidea ( H. bupleuroides group) also differed from the rest of the sample. Hybridogenous groups (“gymh” and “vilh”) shared the same cluster, together with H. naegelianum (sect. Naegeliana), which had been significantly separated from other groups in the UPGMA cluster of previously investigated phenolic compounds (Milutinovi ć et al., 2018). Inclusion of H. gymnocephalum group in sect. Pannosa (“pan” and “wal” cluster) was also not supported.
In this work, proline-SL conjugates in general, as well as known crepiside E (4) were identified in Hieracium genus and generally in whole subtribe Hieraciinae for the first time. Crepiside E (4) was previously identified in only six other Compositae taxa belonging to the genera Crepis L. ( Barda and Skaltsa, 2017), Youngia Cass. ( Lee et al., 2015; Miyase et al., 1985), Prenanthes L. ( Miyase et al., 1987), Lapsana L. ( Fontanel et al., 1999), Ainsliaea DC ( Wu et al., 2011) and Elephantopus L. ( Hisham et al., 1992).
No known copyright restrictions apply. See Agosti, D., Egloff, W., 2009. Taxonomic information exchange and copyright: the Plazi approach. BMC Research Notes 2009, 2:53 for further explanation.