Diphtherophora, de Man, 1880
|
publication ID |
https://doi.org/ 10.11646/zootaxa.4851.2.10 |
|
publication LSID |
lsid:zoobank.org:pub:43C5F7B3-1C03-4738-84CC-FF17B4FE0889 |
|
DOI |
https://doi.org/10.5281/zenodo.4477057 |
|
persistent identifier |
https://treatment.plazi.org/id/924E87E3-9646-4250-FF4C-3FCD1829F70F |
|
treatment provided by |
Plazi |
|
scientific name |
Diphtherophora |
| status |
|
Diphtherophora species phylogeny
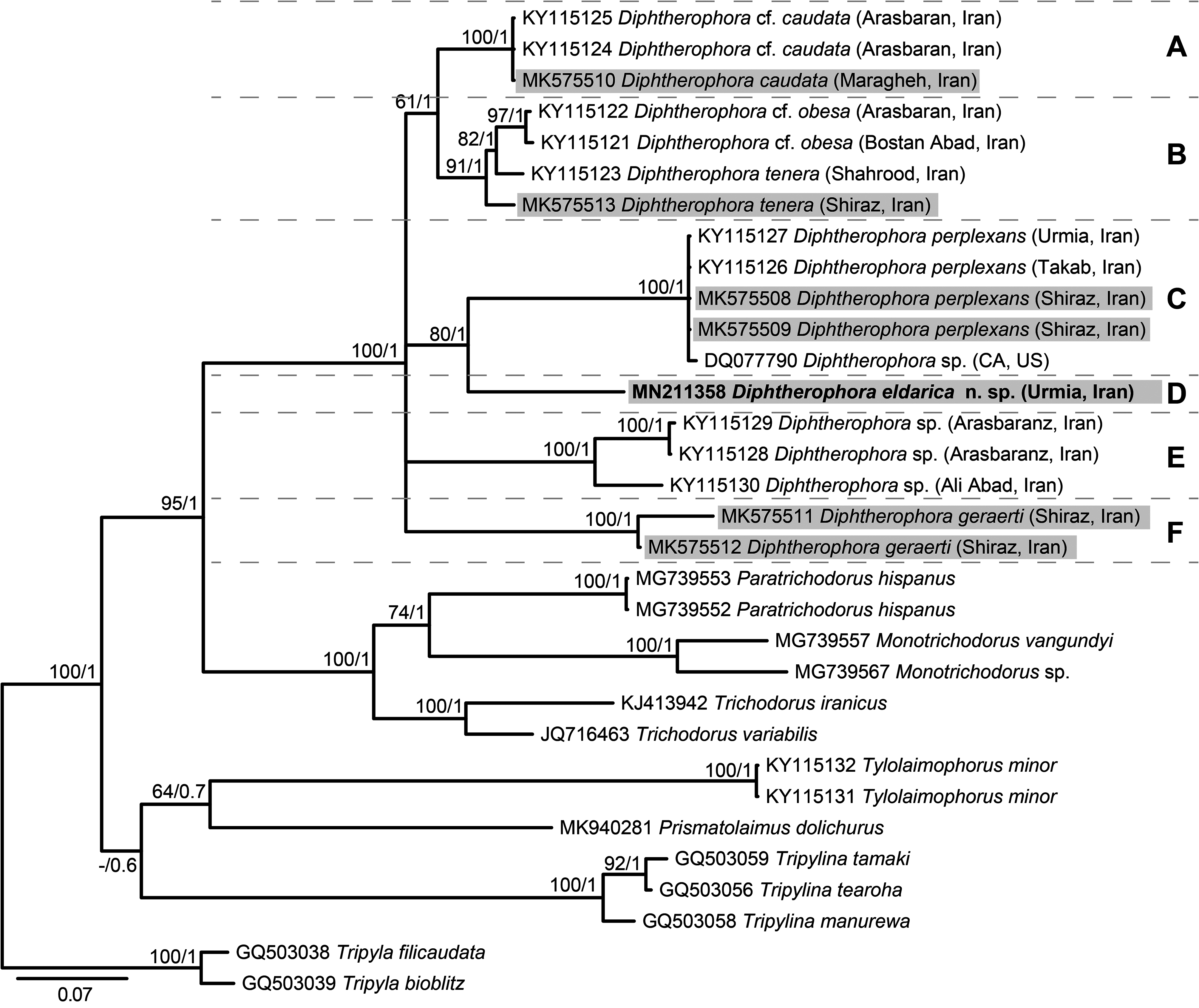
In total, 26 DNA sequences, specifically covering the D2–D3 domains of the 28S rRNA gene, were used in our molecular phylogenetic analyses: 18 Diphtherophora , two Tylolaimophorus , one representative of Prismatolaimidae , six representatives of Trichodoridae , two representatives of Tripylidae , and three representatives of Trischistomatidae . Our final alignment was 785 bp long and contained 326 conserved sites, 455 variable sites, 415 parsimonyinformative sites, and 40 singletons. Phylogenetic analyses based on both inference methods (i.e., ML and BI) were highly congruent with respect to the monophyly of the genus Diphtherophora , the relationships among Diphtherophora species and outgroups, and the phylogenetic placement of D. eldarica n. sp. ( Fig. 4 View FIGURE 4 ).
Phylogenetic trees showed the existence of six strongly supported clades (A–F) within the Diphtherophora group: clade A is represented by D. cf. caudata from Arasbaran (Northwestern Iran) and D. caudata from Maragheh (Northwestern Iran), the latter species recovered in this study; clade B included sequences representing D. cf. obesa and D. tenera from different regions of Northern and Southern Iran, the latter species, however, being paraphyletic; clade C was comprised of D. perplexans from different sites in Northwestern (Takab and Urmia) and Southern Iran (Shiraz), the latter population recovered in this study, and a specimen characterized only as Diphtherophora sp. from CA, USA; clade D was solely represented by the new species D. eldarica n. sp.; clade E harbored uncharacterized Diphtherophora species from different sites in Northern and Northwestern Iran including Arasbaran and Ali Abad; clade F is solely represented by sequences of D. geraerti from Shiraz (Southern Iran). Our phylogenetic analyses strongly supported (ML = 97, BI = 1.0) the new species D. eldarica n. sp. as a sister taxon of clade D, that is, D. perplexans + Diphtherophora sp. ( Fig.4 View FIGURE 4 ).
Genetic divergence (p-distance %) within Diphtherophora clades was relatively low, ranging from 0% (clade A, among D. caudata sequences) to 5.2% (variation within clade E, represented by Diphtherophora sp. sequences from Iran; Table 3 View TABLE 3 ). On the other hand, genetic divergence between Diphtherophora clades was much higher, ranging from 8.2% (clade A vs. B) to as high as 23.7% (clade C vs. F). For comparison, genetic divergence between any of the Diphtherophora clades and Tylolaimophorus (also considered a representative of Diphtherophoridae ) ranged from 25.6% ( Tylolaimophorus vs. clade A) to 32.7% ( Tylolaimophorus vs. clade F). Similarly, genetic divergence between Diphtherophora clades and trichodorid genera were in the same range (24.1% to 30.1%) as those observed for comparisons with Tylolaimophorus . This pattern is also consistent when comparing the genetic divergence between Diphtherophora clades and other outgroup taxa including genera Prismatolaimus de Man, 1880 , Tripyla Bastian, 1865 , and Tripylina Brzeski, 1963 ( Table 3 View TABLE 3 ).
With respect to the new species D. eldarica n. sp., genetic divergence ranged from 13.5% (between clade B, D. tenera + D. cf. obesa ) to 19.5% (between clade E, Diphtherophora sp.). Although all six clades were strongly supported in our analyses, with ML branch support always greater than 98% and BPP always with maximum support, some relationships among Diphtherophora clades (i.e. some of the deeper nodes) were either weakly supported or not recovered by one of the inference methods ( Fig. 4 View FIGURE 4 ).
No known copyright restrictions apply. See Agosti, D., Egloff, W., 2009. Taxonomic information exchange and copyright: the Plazi approach. BMC Research Notes 2009, 2:53 for further explanation.