Plenodomus dezfulensis M. Mehrabi-Koushki, A. Safi & R. Farokhinejad, 2021
|
publication ID |
https://doi.org/ 10.11646/phytotaxa.523.2.2 |
|
DOI |
https://doi.org/10.5281/zenodo.5592618 |
|
persistent identifier |
https://treatment.plazi.org/id/C45BC820-FFD3-2943-71CF-FCEEB3C2FEA1 |
|
treatment provided by |
Plazi |
|
scientific name |
Plenodomus dezfulensis M. Mehrabi-Koushki, A. Safi & R. Farokhinejad |
| status |
sp. nov. |
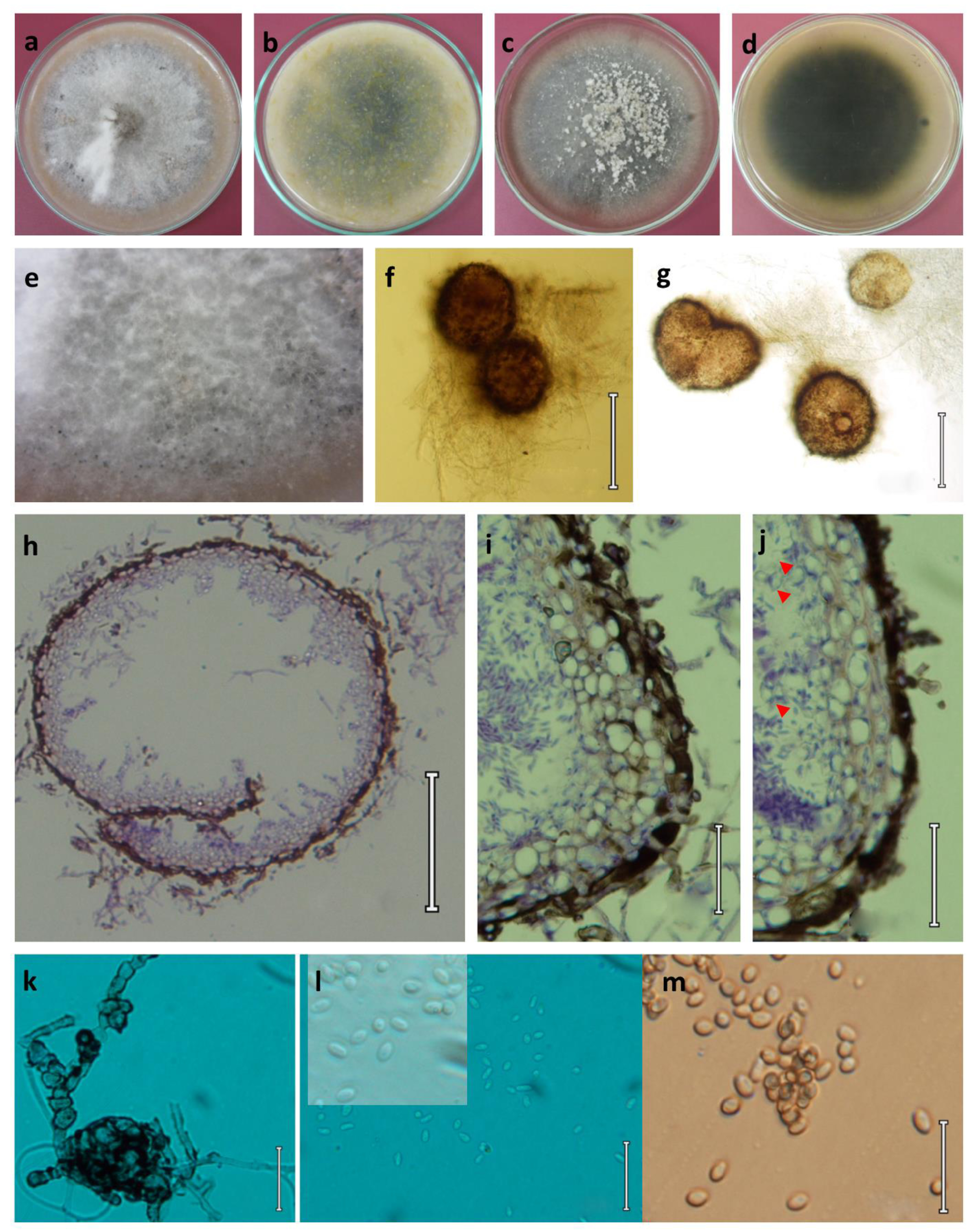
Plenodomus dezfulensis M. Mehrabi-Koushki, A. Safi & R. Farokhinejad View in CoL , sp. nov. ( Fig. 3 View FIGURE 3 )
MycoBank: MB 839660
Holotype: IRAN. Khuzestan Province; Dezful (Safiabad), isolated from leaf spot of Brassica napus subsp. napus (Brassicaceae) , December 2019, A. Safi (holotype, IRAN 18084F ; ex-type cultures, IRAN 4159C = SCUA-Ahm-S41).
Etymology: Species epithet refers to Dezful County in Khuzestan Province ( Iran), where the fungus was collected.
Asexual morph: Coelomycetous. Conidiomata pycnidial, scattered and irregular, solitary or confluent, partly submerged in the agar, globose to subglobose, with hyphal outgrowths, ostiolate, papillate, brown to tan brown with a darker border, thick-walled, (89–)102–271.4 56.3–238(–263.7) µm, 95% confidence limits = 158.1–182.2 109.8– 129.7 µm, (± SD = 170.1 ± 46.6 119.8 ± 38.6 µm, n= 60). Pycnidial wall variable in thickness, composed of several layers with thick-walled cells of textura angularis, somewhat elongated cells at the base of the pycnidia. Ostioles slightly papillate, with a narrow pore or opening via a rupture. Conidiogenous cells hyaline, smooth, ovoid to doliiform, 3.5–5.2 2.5–4 µm. Conidia ellipsoidal to oblong, rarely allantoid, straight, sometime very slightly curved, hyaline but greyish brown in mass, guttulate near each end, rounded at both ends, aseptate, 3.15–7.4 1– 2.2(–2.7) µm, 95% confidence limits = 4.2–4.7 1.9–2.1 µm, (± SD = 4.5 ± 0.8 1.9 ± 0.3 µm, n= 60). Chlamydospores occasionally found in old cultures, unicellular or occasionally multicellular (pseudosclerotioid and dictyosporous). Swollen cells terminal or intercalary, solitary or in clusters. Sexual morph: not observed.
Culture characteristics: colonies on PDA attaining 40–45 mm diam. after 8 days of incubation at 25 ± 0.5 °C and 52–55 mm diam. at 30 ± 0.5 °C, circular with filiform margin, initially greenish grey to olivaceous grey, with age becoming darker, finally grey, floccose, in some area covered by dirty-white, woolly mycelial masses; reverse brownish green with a grey margin in young colonies, blackish grey in old colonies. Colonies on OA attaining 43–47 mm diam. after 8 days of incubation at 25 ± 0.5 °C and 47–50 mm diam. at 30 ± 0.5 °C, circular with filiform margin, white to dark white, with age becoming grey, cottony and covered by white aerial mycelial, pycnidia scattered in colony margin as black dots; reverse grey.
Additional specimen examined: IRAN. Khuzestan Province; Dezful, isolated from leaf spot of Brassica napus subsp. napus (Brassicaceae) , January 2020, A. Safi (SCUA-Ahm-S41-2).
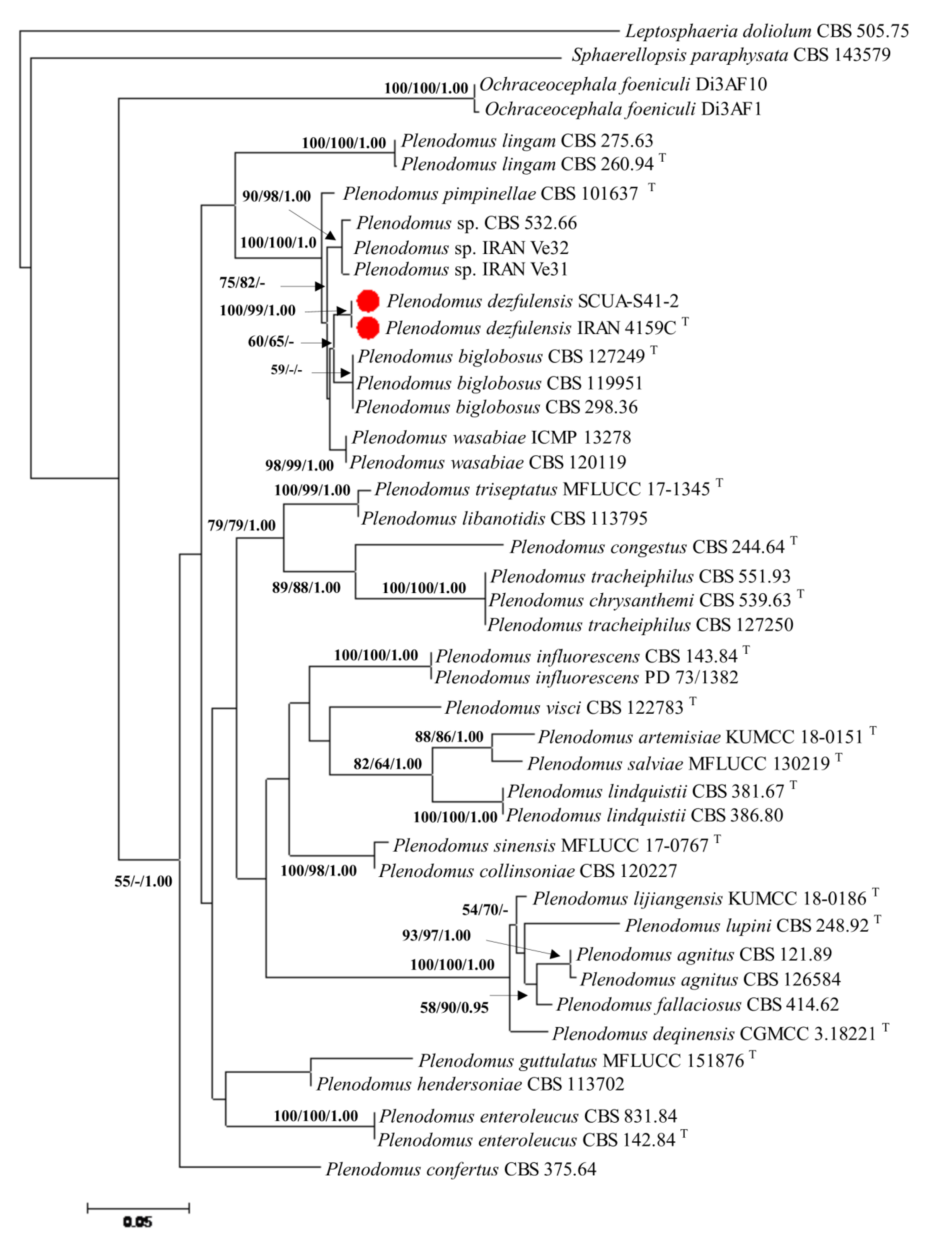
Note: Plenodomus dezfulensis formed a sister lineage with P. biglobosus ( Fig. 2 View FIGURE 2 ), but it can easily be distinguished from this species by pycnidia having hyphal outgrowths and lacking cylindrical neck ( Shoemaker & Brun 2001). Single nucleotide polymorphisms (SNP) analysis of three regions showed the new species and closest strain of P. biglobosus (CBS 119951) had 2 base pair difference (0.46%) across 435 nucleotides of the ITS region, 7 different base pairs (2.3%) across 306 nucleotides of the tub2 region, and a difference of 16 base pairs (2.2%) across 712 nucleotides of the rpb2 region. Two other closely related species of the new species are P. pimpinellae (Lowen & Sivan.) Gruyter, Aveskamp & Verkley , and P. wasabiae . Plenodomus dezfulensis can be easily distinguished from P. pimpinellae by narrower, in culture formed pycnidia lacking long elongated neck ( Boerema et al. 2004). In the comparison of DNA sequences of two regions (ITS: 435 bp, tub2: 306 bp), Plenodomus dezfulensis differs from P. pimpinellae in 13 base pairs (1.8%, ITS: 3 bp, tub2: 10 bp) and from P. wasabiae in 12 base pairs (1.6%, ITS: 4 bp, tub2: 8 bp). The rpb2 data of both species are not available for the comparison.
| A |
Harvard University - Arnold Arboretum |
No known copyright restrictions apply. See Agosti, D., Egloff, W., 2009. Taxonomic information exchange and copyright: the Plazi approach. BMC Research Notes 2009, 2:53 for further explanation.