Camellia gymnogyna
|
publication ID |
https://doi.org/ 10.1016/j.phytochem.2022.113167 |
|
persistent identifier |
https://treatment.plazi.org/id/CE1E87E4-475A-FFF3-FFAF-F8F342BEF919 |
|
treatment provided by |
Felipe |
|
scientific name |
Camellia gymnogyna |
| status |
|
2.3. Cloning of candidate N-methyltransferase genes from C. gymnogyna View in CoL View at ENA
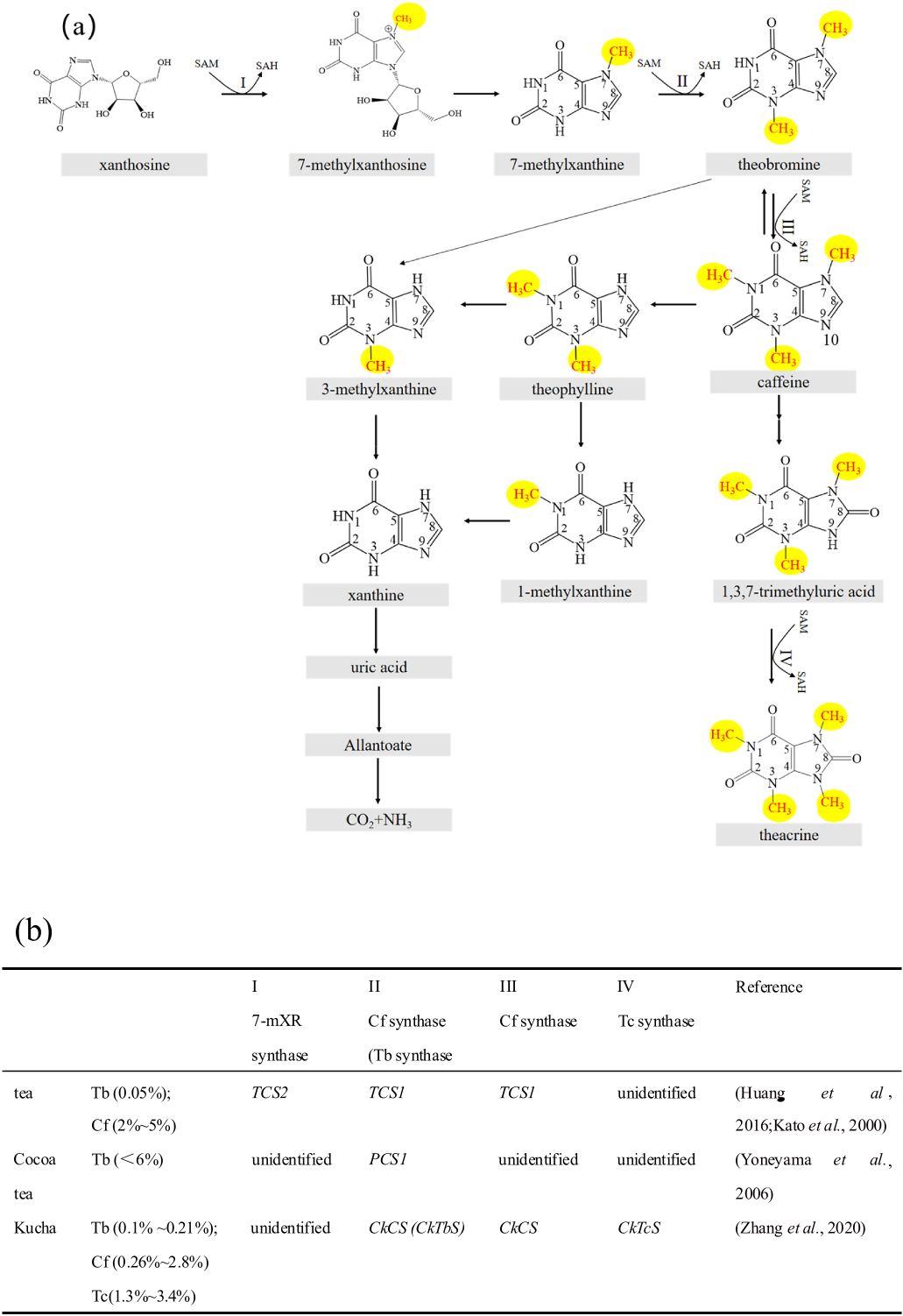
According to the blast analysis of the transcriptome data of C. gymnogyna was conducted using the amino acid sequence of tea Cf synthase (TCS1), two candidate N -methyltransferases were identified. Based on their sequence information, we designed two different primers to clone the candidate N -methyltransferase genes by polymerase chain reaction (PCR). Two cloned N -methyltransferase genes were named GCS1 and GCS2. In addition, according to the sequence information of the Tb synthase gene reported previously ( Teng et al., 2019), we designed the primers to clone a N -methyltransferase gene and renamed it GCS3.
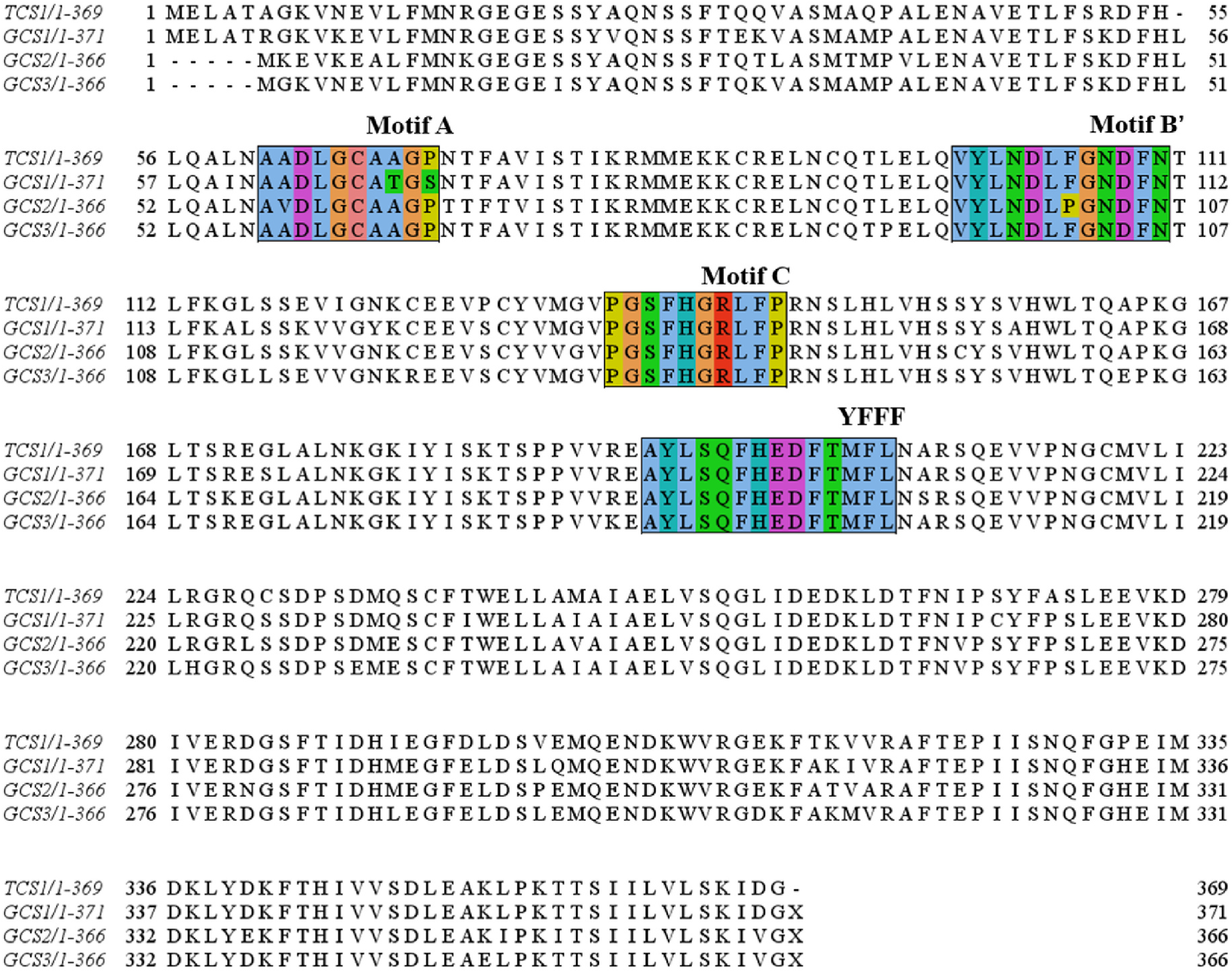
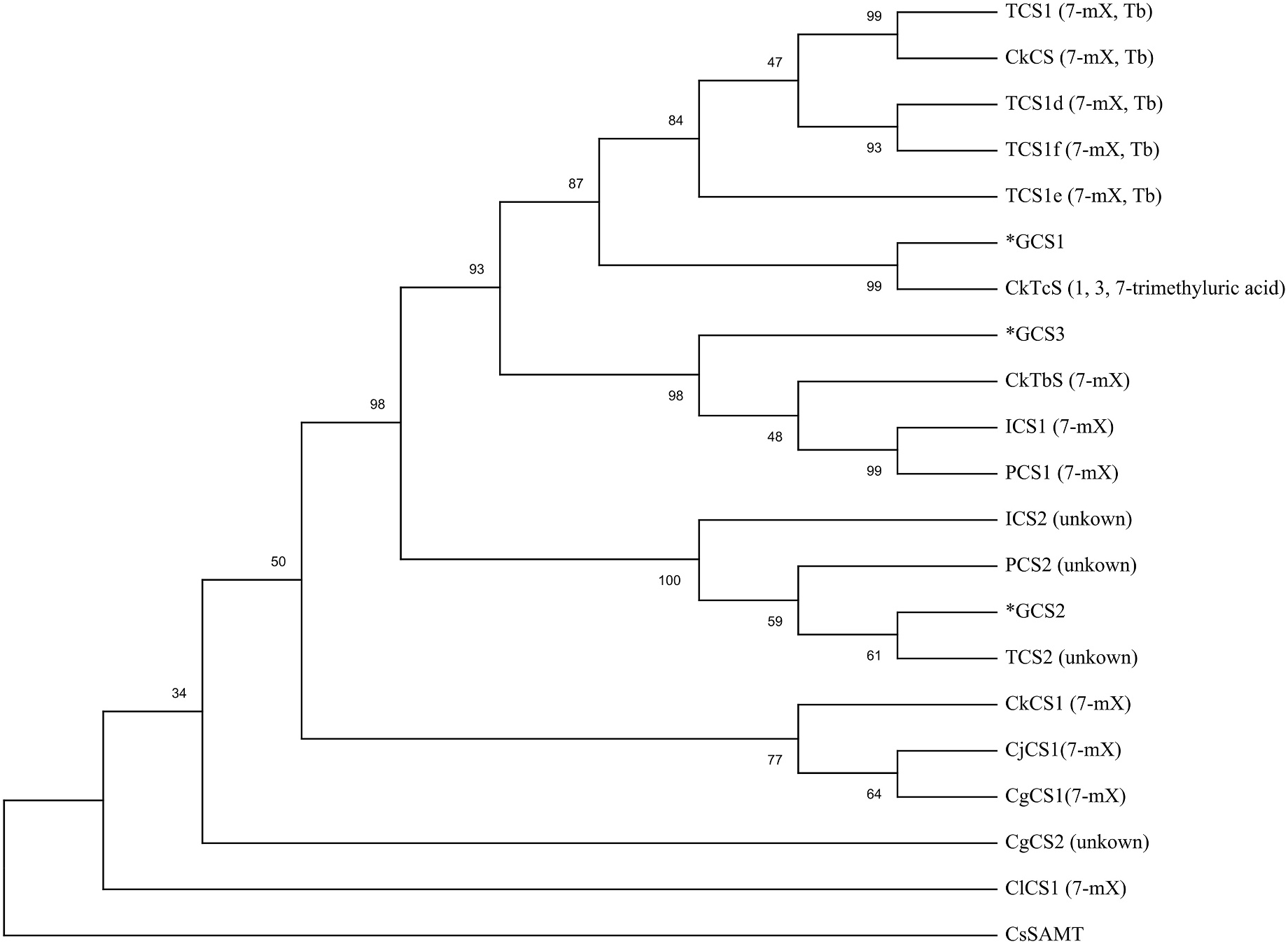
GCS1 consisted of 1113 bp and encoded 370 amino acid residues. Both GCS2 and GCS3 were composed of 1098 bp and encoded 365 amino acids. GCS1, GCS2, and GCS3 shared a high degree of sequence similarity with N -methyltransferase genes from other Camellia plants ( Table 2). The amino acid sequence of GCS1 showed the highest homology with the Tc synthase gene CkTcS (97.0%), while the amino acid sequence of GCS2 shared highest homology with TCS2 (98.6%). The amino acid sequence of GCS3 was more than 97% homologous with CkTbs and ICS1. Sequence alignments revealed that GCS1, GCS2, and GCS3 contained typical motif A, motif B ′, motif C, and YFFF region ( Fig. 4 View Fig ). Phylogenetic analysis indicated that GCS1 was more closely related to CkTcS, GCS2 was more closely related to TCS2, and GCS3 was more closely related to Tb synthases (CkTbS, ICS1 and PCS1) ( Fig. 5 View Fig ). Interestingly, Tc synthase was more closely related to Cf synthase than other N -methyltransferases.
2.4. Biochemical Characterization of GCS1, GCS2 and GCS3
GCS1, GCS2, and GCS3 were expressed in Escherichia coli , and the molecular masses of their proteins were 41.6 kDa, 40.8 kDa and 41.0 kDa, respectively. The soluble expressions of heterologous proteins in Escherichia coli remain a serious bottleneck in protein production ( Esposito and Chatterjee, 2006). To obtain more soluble
GCS1/GCS2/GCS3 proteins, we explored concentrations of the inducer. When the induction temperature is 37 ◦ C, the total amount of GCS1/GCS2/GCS3 proteins was the highest, while the contents of soluble proteins was lowest. The solubility of GCS1/GCS2/GCS3 proteins were slightly increased with lower expression temperatures. When the concentration of isopropyl β- D-1-thiogalactopyranoside (IPTG) was 0.1 mM, the content of soluble proteins was the highest.
The incubation of GCS1 crude enzyme extracts with 1,3,7-trimethyluric acid substrates led to the formation of a new peak (Supplement Figure 1 View Fig and Supplement Figure 3 View Fig ) with the same retention time and absorption spectrum as Tc. The incubation of GCS3 crude enzyme extracts with 7-mX substrates led to the formation of a new product peak (Supplement Figure 2 View Fig and Supplement Figure 4 View Fig ) with the same retention time and absorption spectrum as Tb. The results indicated that GCS1 catalyzed conversion of 1,3,7-trimethyluric acid to Tc, and GCS3 catalyzed conversion of 7-mX to Tb. Dichloromethane extraction of GCS1 and GCS3 crude enzyme mixture showed few impurity peaks, indicating its better efficacy than ethyl acetate extraction. The GCS2 had no methyltransferase activity using X derivatives as substrate.
No known copyright restrictions apply. See Agosti, D., Egloff, W., 2009. Taxonomic information exchange and copyright: the Plazi approach. BMC Research Notes 2009, 2:53 for further explanation.
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |