Myxobolus arariensis, Abrunhosa & Sindeaux-Neto & Santos & Hamoy & Matos, 2018
|
publication ID |
https://doi.org/ 10.11646/zootaxa.4482.1.8 |
|
publication LSID |
lsid:zoobank.org:pub:C52A9D11-88C9-46BA-917E-3940F3C8A3A3 |
|
DOI |
https://doi.org/10.5281/zenodo.5980187 |
|
persistent identifier |
https://treatment.plazi.org/id/DF4E4F37-B034-FFBC-D4AE-995CDB27F9C6 |
|
treatment provided by |
Plazi |
|
scientific name |
Myxobolus arariensis |
| status |
sp. nov. |
Myxobolus arariensis n. sp.
( Figs 1–3 View FIGURE 1 View FIGURE 2 View FIGURE 3 )
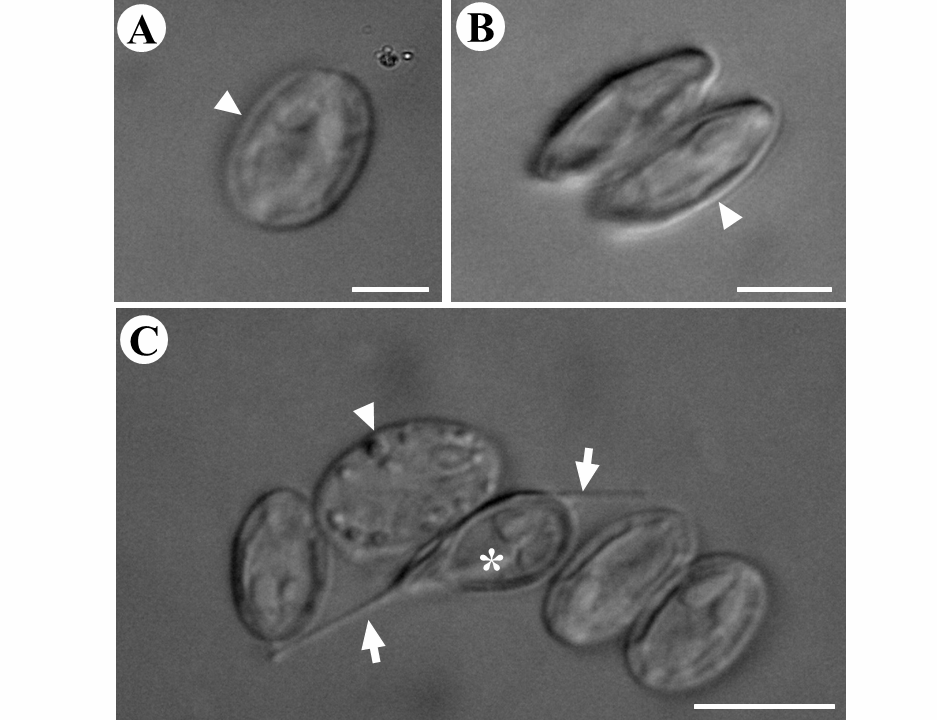
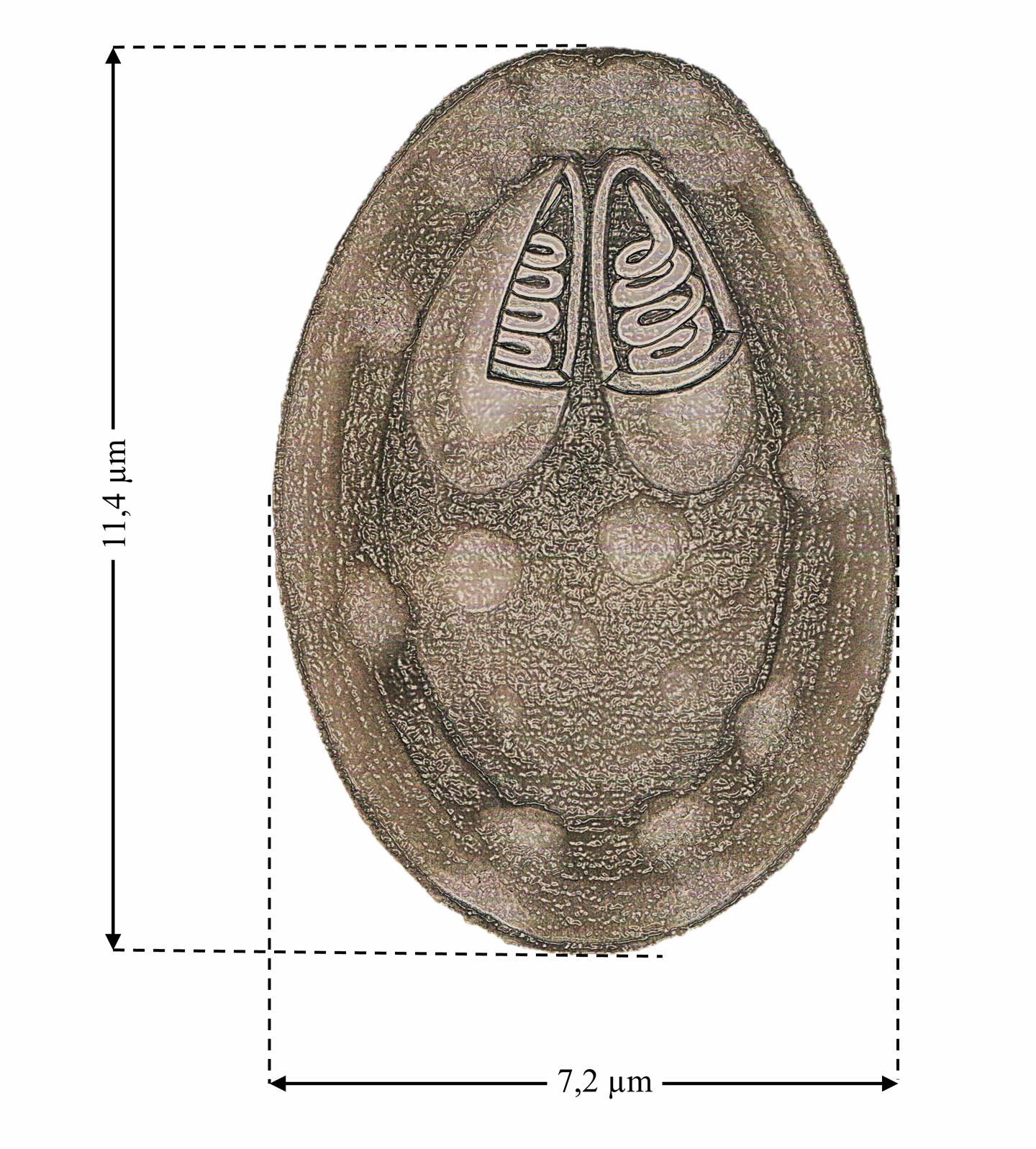
Morphological description. Mature spores are ellipsoidal in shape, with a mean length of 11.4 µm (10.7– 12.6) and mean width of 7.2 µm (6.4–7.9). Each spore contains two polar capsules (PCs) of equal size 4.0 ± 0.7 µm (3.6–4.3) long and 1.9 ± 0.36 µm (1.7–2.2) in width ( Figs. 1B View FIGURE 1 and 3 View FIGURE 3 ) ( Table 1).
Type host. Rhamdia quelen (Quoy & Gaimard 1824)
Site of infection. epaxial and hypaxial layers of the musculature, with plasmodia containing numerous spores.
Type-locality. Arari River , Cachoeira do Arari on Marajó Island, northern Brazil.
Etymology. The species was named for the locality of origin, the Arari River in northern Brazil.
Specimens deposited. Microscope slides containing spores from the muscle layer, prepared using the paraffin technique, stained in Gutierrez and mounted in Entellan were deposited in the International
Protozoan Type Collection of the National Institute for Amazonian Research ( INPA) in Manaus , Amazonas state, Brazil (catalog number: INPA /027). The partial 18S rDNA sequence was deposited in GenBank under accession number MG572219 View Materials .
Prevalence. Three of twenty-five R. quelen examined 12% (3/25) had plasmodia of an unknown parasite from the genus Myxobolus .
Remarks. M. arariensis can be differentiated morphologically from all seven Myxobolus species known to infect the muscle tissue of freshwater fish ( Table 1). The new species can be distinguished from M. tasikkenyirensis (Székely et al. 2009a) and M. groenlandicus ( Buchmann et al. 2012) by the different shape of the anterior extremity of the spores, and from M. leptobarbi (Székely et al. 2009b) by the same trait. The length of M. arariensis (11.4 µm) is most similar to that of M. lentisuturalis (Dyková et al. 2002) , which is 11.8 µm long, whereas M. omari (Székely et al. 2009b) is the shortest species, at 7.9 µm. Anomalous spores with a caudal filament and lack of ornamentation on the external wall were also observed ( Fig. 2C View FIGURE 2 ).
Histology. The histological analysis revealed the presence of cysts of M. arariensis lodged in the fibers of the skeletal muscles ( Fig. 4 View FIGURE 4 ). Immature spores were observed in the most external layer of the cyst, with mature spores being found more internally. The cyst wall is thick and fibrous, and the adjacent musculature was compressed, with the sarcoplasm frayed, and evidence of a necrotic reaction caused by this compression.
Molecular data. In the molecular analysis, the specific pair of myxozoan primers (MC5-MC3) amplified 974 bps of the 18S rDNA gene of the spores obtained from the plasmodia found infecting the musculature of R. quelen . The BLAST search of the 18S rDNA sequence data (974 bps) of the Myxobolus species parasitizing R. quelen found no identical myxozoan sequence in GenBank, although a similarity of at least 85% was found with four species: Myxobolus cordeiroi ( KF296353 View Materials , 90% similarity), Myxobolus sp. GA2 ( KU 170935 View Materials , 86%), Myxobolus lentisuturalis ( AY278563 View Materials , 85%), and Myxobolus cultus ( HQ613409 View Materials , 85%).
The optimal evolutionary model for maximum likelihood (ML) and Bayesian analysis were determined by jModelTest 3.0 (Posada, 2008) which identified the best evolutionary model as the general time reversible model (GTR + I + G), using Akaike information criteria. Nucleotide frequencies were estimated from the data (A = 0.2574, C = 0.1848, G = 0.2625, T = 0.2326) and six rates of nucleotide substitution calculated as AC = 0.8659, AG = 2.6388, AT = 1.7658, CG = 0.4883, CT = 3.4814, GT = 1.000. The proportion of invariable site was 0.5565 and the alpha value of gamma distribution parameter 0.3612. Two independent runs were conducted with 4 chains for 2 million generations for Bayesian analysis. Ceratomyxa shasta ( AF001579 View Materials ) e C. amazonensis ( KX236169 View Materials ) was designated as outgroup. Phylogenetic trees were sampled every 100 generation.
Characteristics of some Myxobolus species. Abbreviations: FC = Capsule Formate, SL, spore length, SW = Spore Width, PCL = Polar Capsule Length, PCW= Polar Capsule Width. PC = relative size of the polar capsules (= = equal in size, # = different in size, or equal and different); All measurements are given in micrometers.
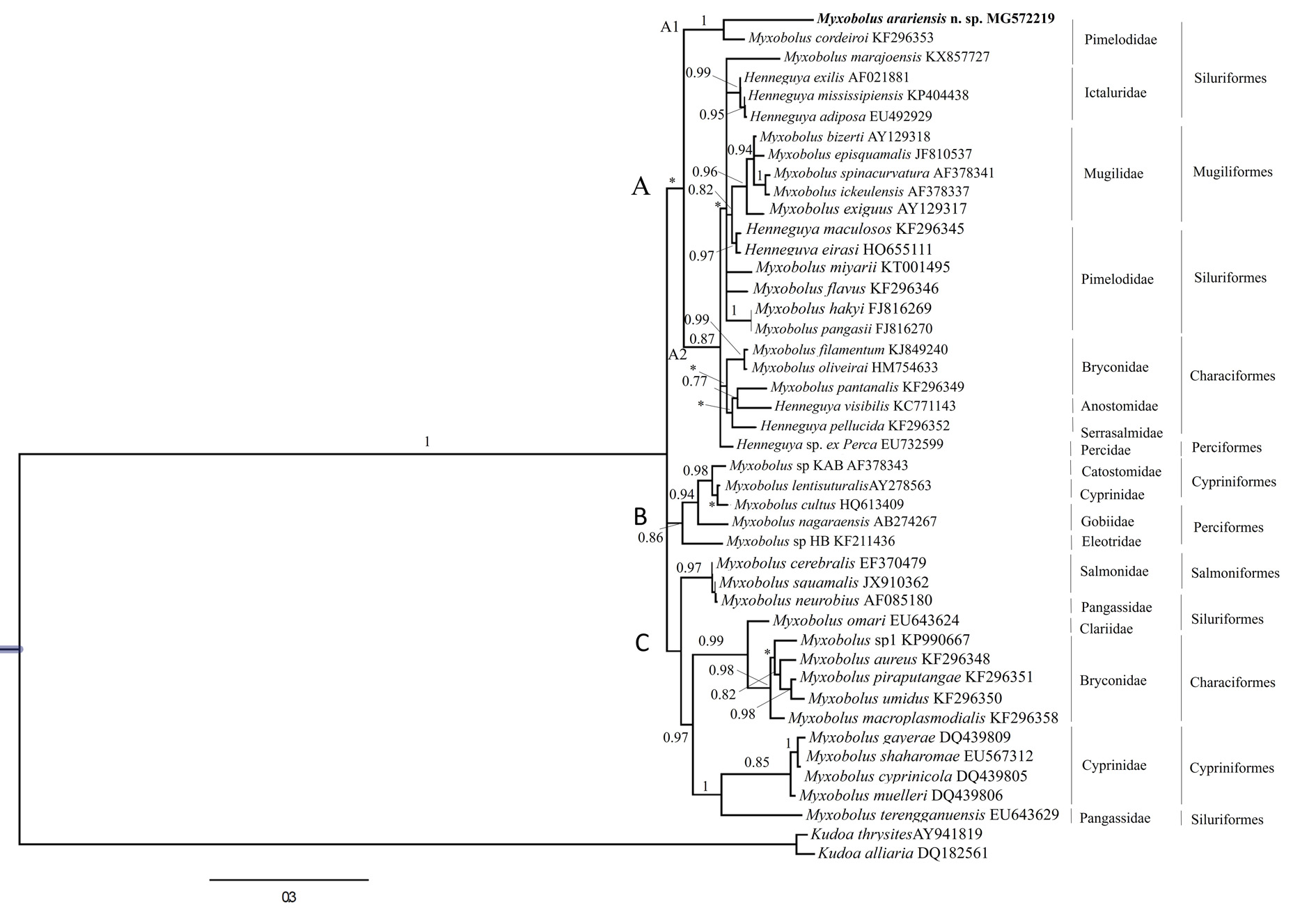
In the phylogenetic analysis, trees generated by Bayesian Inference (BI) had similar topologies, but with different support values at some nodes. A strong clustering tendency was found according to phylogenetic affinities. The phylogram indicated the existence of three clades, A, B and C the first paraphyletic, includes species of Henneguya and Myxobolus formed by the freshwater and marine water ( Mugiliformes ).
The clade A subdivide into 2 subclades, A1 and A2. The subclade A1 shows M. arariensis grouping on the same branch with M. cordeiroi and with adjacent subclade with M. marajoensis species, having same host. The other subclade, A2 have the presence of the Myxobolus and Henneguya , corroborating the characteristic of the Myxobolus genus to be paraphyletic. On the other hand, the clades B and C have agrouped species of various Myxobolus parasites of freshwater fish belonging to same and different Order ( Table 3).
The type of host defines a well-supported freshwater and marine water clade of Myxobolus and Henneguya (clade A) that infect fish of the orders Siluriformes , Mugiliformes , Characiformes and Perciformes . However, M. arariensis , which infects silurids, evolved independently from the Myxobolus subclade A1 that infects fish of the families Pimelodidae and Ictaluridae . The tree presented a similar topology for the other clades, clustering according to the taxonomic order of the host. The clade B groups Myxobolus , of the orders Cypriniformes and Perciformes . The clade C is also compounded by Myxobolus groups that parasitize hosts of the different orders Salmoniformes , Siluriformes , Characiformes and Cypriniformes ( Fig. 5 View FIGURE 5 ).
The p distance found between M. arariensis and any other Myxobolus species that had Siluriformes as host ranged from 11.6 to 22.1% ( Table 2), which reinforces the definition of M. arariensis as a new species. The myxosporidian sequences analysed are showing in the Table 3.
……continued on the next page
| INPA |
Instituto Nacional de Pesquisas da Amazonia |
No known copyright restrictions apply. See Agosti, D., Egloff, W., 2009. Taxonomic information exchange and copyright: the Plazi approach. BMC Research Notes 2009, 2:53 for further explanation.