Pseudochactas ovchinnikovi, Gromov 1998 LP, 2303
|
publication ID |
https://doi.org/ 10.1093/isd/ixac028 |
|
persistent identifier |
https://treatment.plazi.org/id/4C167777-FFF3-2926-5F63-89A9FAEC5765 |
|
treatment provided by |
Felipe |
|
scientific name |
Pseudochactas ovchinnikovi |
| status |
|
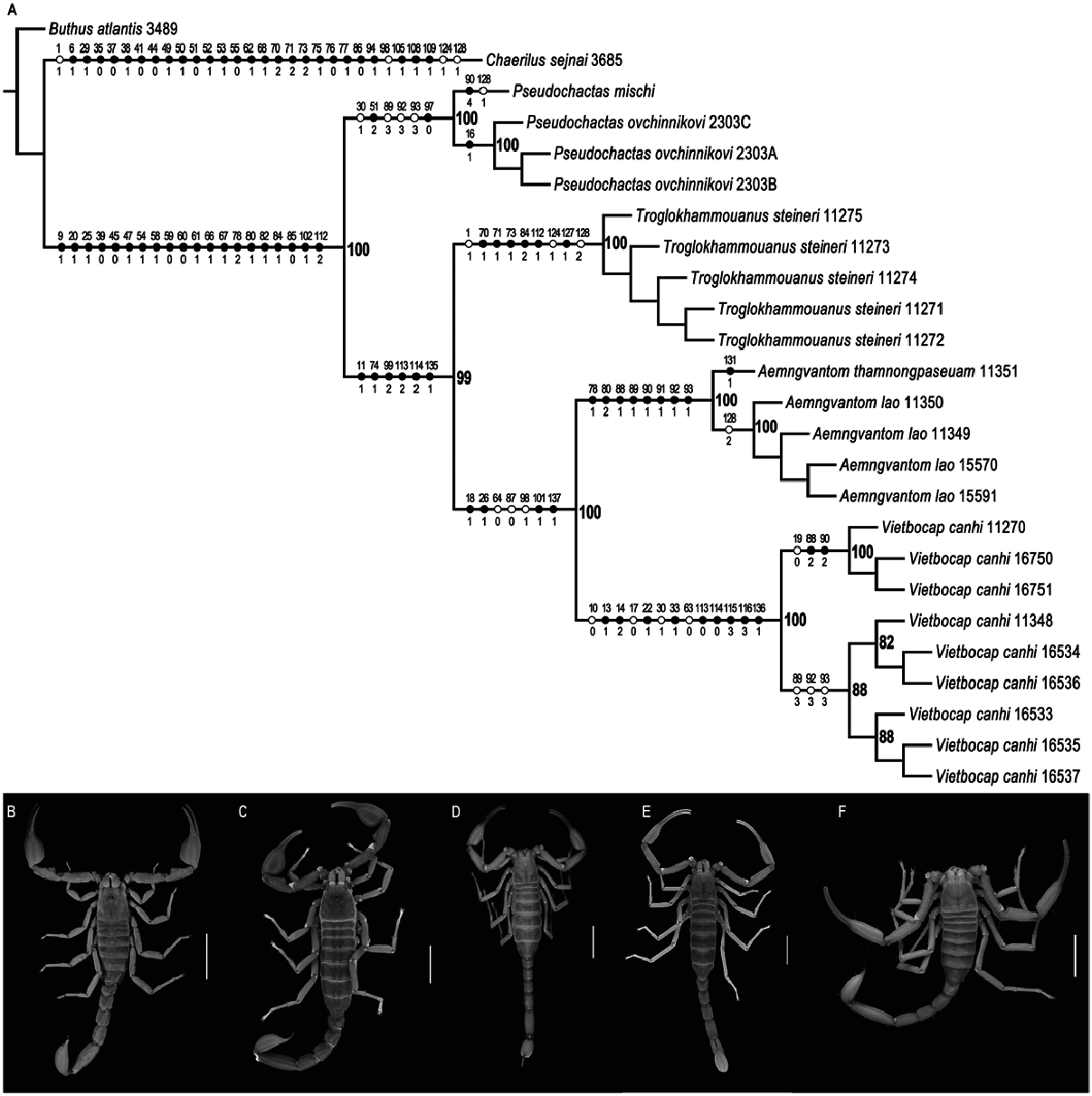
P. ovchinnikovi View in CoL (68%), Pseudochactas (58%), the Thiên Ðu lation of V. canhi (28%), and the Southeast Asian clade comprising Troglokhammouaninae Prendini et al. 2021 and Vietbocapinae. Jackknife support values in the separate analysis of the morphological dataset with parsimony and equal weighting were also high (≥ 79%) except for P. ovchinnikovi (70%), A. lao (66%), the Thiên Ðu population of V. canhi (54%) and intraspecific relationships among A. lao , P. ovchinnikovi , T. steineri and V. canhi (> 10%).
Simultaneous analyses of the morphological and molecular datasets with BI, ML and parsimony with equal or implied weighting (k = 1, 3, 10, 60, 100) were congruent with separate analyses of the molecular and morphological datasets and recovered all subfamilies, genera, species and populations as monophyletic. Posterior probability and bootstrap support values obtained by simultaneous analysis of the morphological and molecular datasets were generally high in the analyses with BI (≥0.99), ML (≥99%), and parsimony with equal weighting (≥ 100%). Jackknife support values were also high (≥83%) in the parsimony analysis with equal weighting except for the following, which received low to moderate support: P. ovchinnikovi (63%) and intraspecific relationships among V. canhi and T. steineri (>10%). Unambiguous morphological synapomorphies were optimized on the ML tree obtained by simultaneous analysis of the morphological and molecular datasets, the preferred phylogeny ( Fig. 2 View Fig ). Output files from phylogenetic analyses are publicly available in figshare: 10.6084/m9.figshare.21067528.
No known copyright restrictions apply. See Agosti, D., Egloff, W., 2009. Taxonomic information exchange and copyright: the Plazi approach. BMC Research Notes 2009, 2:53 for further explanation.
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |