Conocephalum salebrosum, Szweyk., Buczk. & Odrzyk
|
publication ID |
https://doi.org/ 10.1016/j.phytochem.2019.04.013 |
|
DOI |
https://doi.org/10.5281/zenodo.10580632 |
|
persistent identifier |
https://treatment.plazi.org/id/03AB0102-996D-E01B-AB4B-08D9FCF93A23 |
|
treatment provided by |
Felipe |
|
scientific name |
Conocephalum salebrosum |
| status |
|
2.1. Production of axenic culture of C. salebrosum View in CoL and chemical profiling
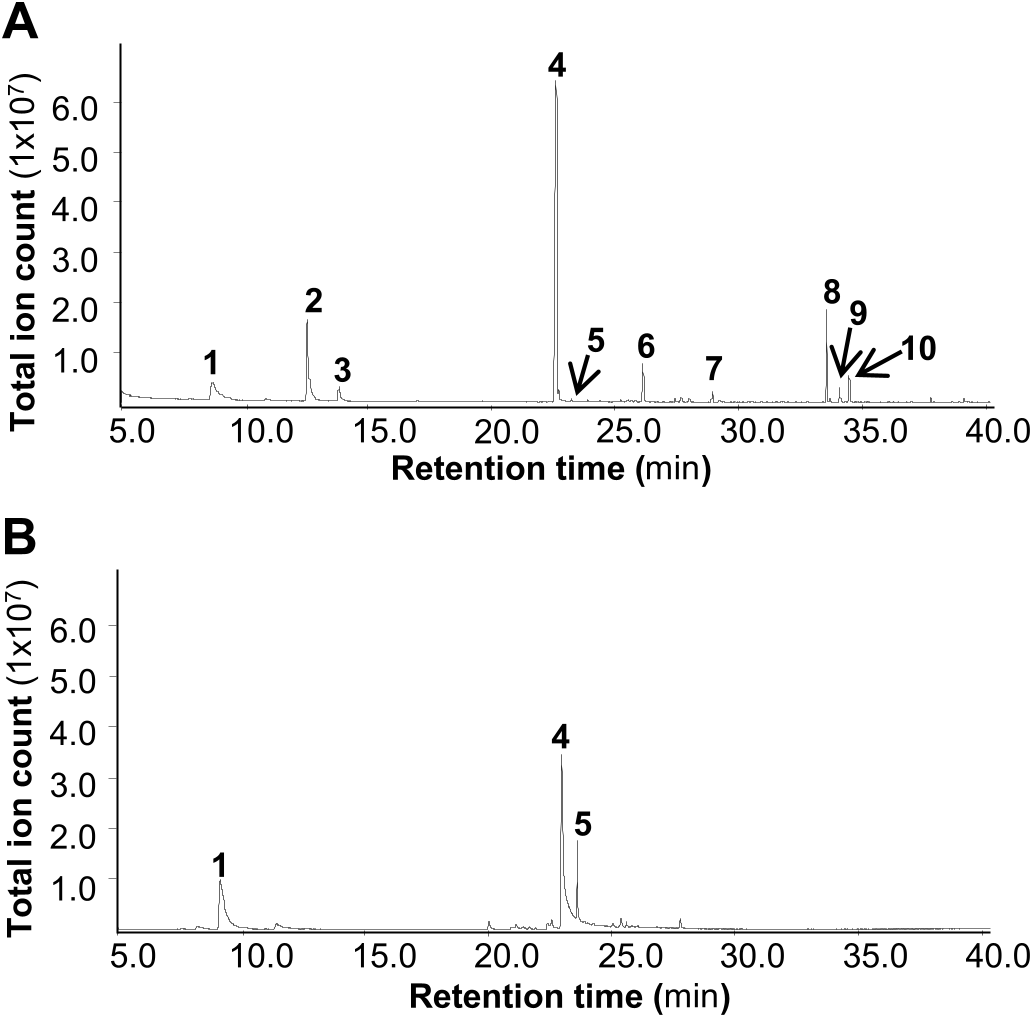
The sample collected from Illinois (IL) morphologically matched C. salebrosum , a segregate species of C. conicum ( Szweykowski et al., 2005) . This population is the chemotype with methyl (E)-cinnamate as the dominant compound described by Wood et al. (1996), and was therefore chosen as the model chemotype for most of the experiments in this study. Live plants were collected in the field and grown in axenic culture. Chemical profiling analysis was performed using gas chromatography–mass spectrometry (GC-MS) to determine the volatile components in the axenically grown gametophytes of this species of the C. conicum complex. From organic extraction, a total of 10 volatiles were detected ( Fig. 1A View Fig ), of which methyl (E)-cinnamate was the dominant component, accounting for 50.5% of the total volatiles. The monoterpene sabinene was the second most abundant with a concentration of 20% of that of methyl (E)-cinnamate. The thallus of C. salebrosum , when growing in the wild, possesses a characteristic aromatic odor. To determine whether C. salebrosum grown axenically on culture medium emits volatiles, we performed headspace collection of explants of C. salebrosum , which was analyzed by GC-MS. Three dominant compounds were detected ( Fig. 1B View Fig ). All of them were also detected by organic extraction. Similar to organic extraction, methyl (E)-cinnamate was the most abundant compound in the volatile bouquet of C. salebrosum ( Fig. 1B View Fig ).
2.2. Isolation of SABATH genes from C. salebrosum View in CoL and gene expression analysis
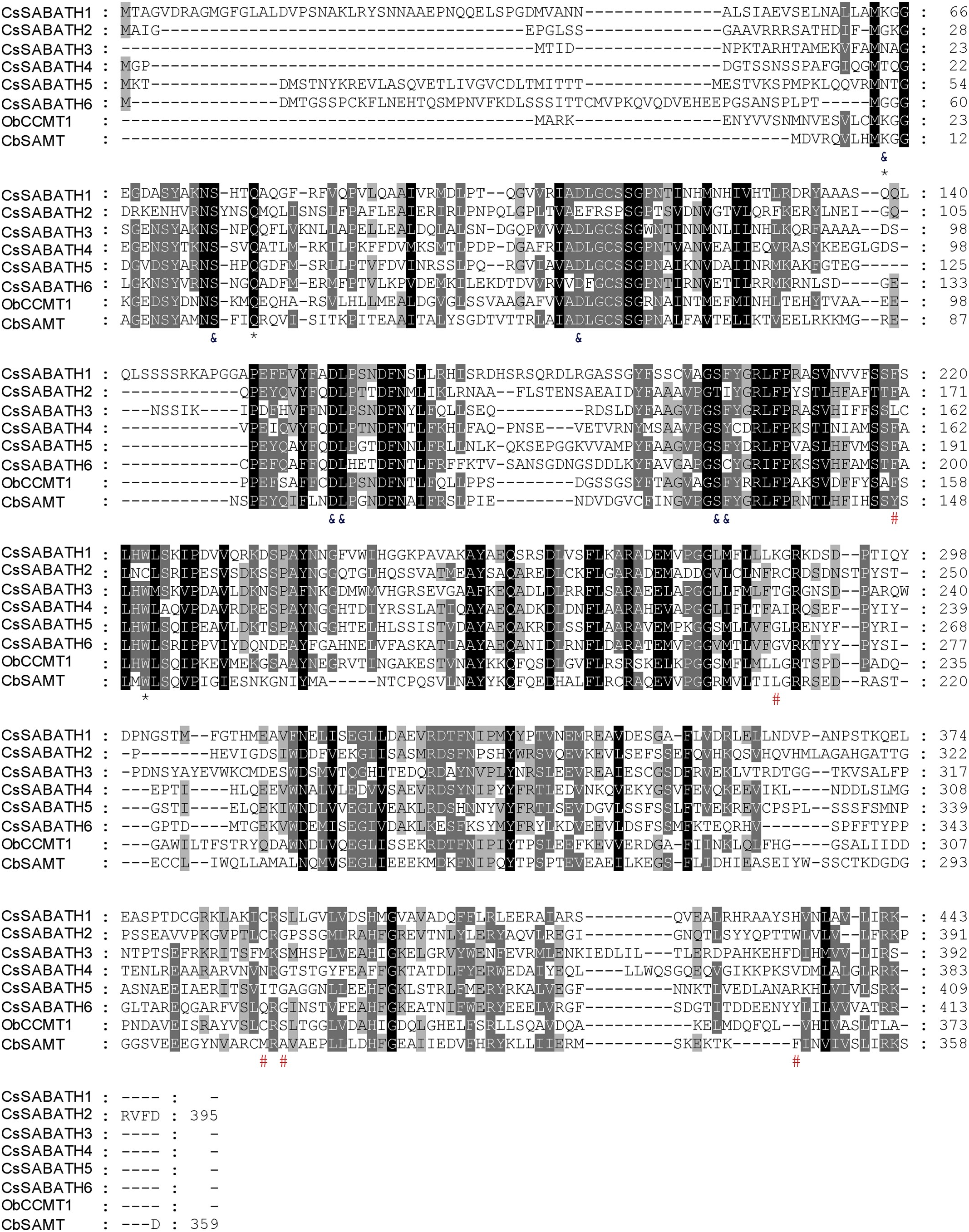
We hypothesized that in the C. salebrosum methyl (E)-cinnamate is synthesized by the action of SABATH methyltransferase. To identify SABATH genes, we analyzed the transcriptome of C. conicum prepared from vegetative thalli (https://sites.google.com/a/ualberta.ca/onekp), which was produced by the 1 KP consortium ( Matasci et al., 2014). Via a HMMR search ( Finn et al., 2011) using Methyltransf_7 Pfam profile ( Finn et al., 2015) as query, a total of nine unigenes encoding SABATH proteins were identified. Six of them appear to be in full-length when compared to other SABATH proteins and their corresponding orthologs in C. salebrosum were designated CsSABATH1-6 ( Table S1 View Table 1 ). Using RT-PCR, full-length cDNAs for all six CsSABATH genes were cloned from the thallus of the axenically grown C. salebrosum . The lengths of six proteins range from 383 to 443 amino acids ( Fig. 2 View Fig ). Sequence similarities among CsSABATHs range from 37% to 52%. Their similarities to ObCCMT1, one known CAMT from basil, are between 36% and 48%. The amino acid residues for binding the methyl donor SAM among CsSABATHs and ObCCMT1 are identical. The residues that interact with the carboxyl moiety of the substrate are also conserved. However, the residues that interact with the aromatic moiety are significantly different ( Fig. 2 View Fig ).
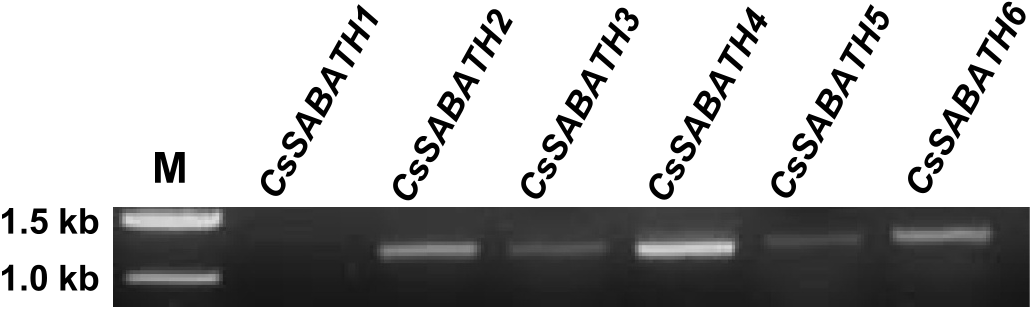
With six SABATH genes cloned from C. salebrosum , next we compared their expression levels using semi-quantitative reverse transcription polymerase chain reaction (RT-PCR) analysis. At 25 cycles of
PCR reactions, transcripts for five of the six genes could be detected from thallus tissue, the exception being CsSABATH1 ( Fig. 3 View Fig ). CsSABATH4 showed the highest level of expression. While CsSABATH2 and CsSABATH6 showed also higher expression levels. CsSABATH3 and CsSABATH5 were found to be expressed at relatively low levels ( Fig. 3 View Fig ).
No known copyright restrictions apply. See Agosti, D., Egloff, W., 2009. Taxonomic information exchange and copyright: the Plazi approach. BMC Research Notes 2009, 2:53 for further explanation.
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |