Nycticeinops happoldorum ( Hutterer et al ., 2019 )
|
publication ID |
https://doi.org/10.3161/15081109ACC2023.25.1.002 |
|
DOI |
https://doi.org/10.5281/zenodo.10261417 |
|
persistent identifier |
https://treatment.plazi.org/id/03860761-0E03-0A4C-9971-FA0C193B95BE |
|
treatment provided by |
Felipe |
|
scientific name |
Nycticeinops happoldorum ( Hutterer et al ., 2019 ) |
| status |
|
Nycticeinops happoldorum ( Hutterer et al., 2019) View in CoL
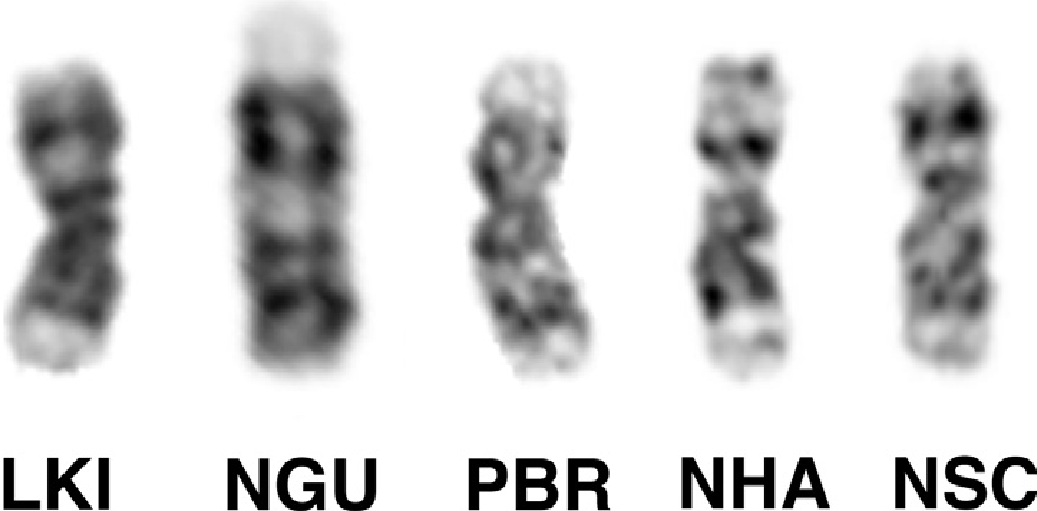
The examined male of N. happoldorum (NHA) showed a karyotype with 2n = 24 chromosomes (FNa = 44) with eight large- to medium-sized meta- to submetacentric autosomal pairs and three medium-sized subtelocentric pairs. The X chromosome was a medium-sized submetacentric and the Y chromosome was a small metacentric chromosome ( Fig. 8 View FIG ).
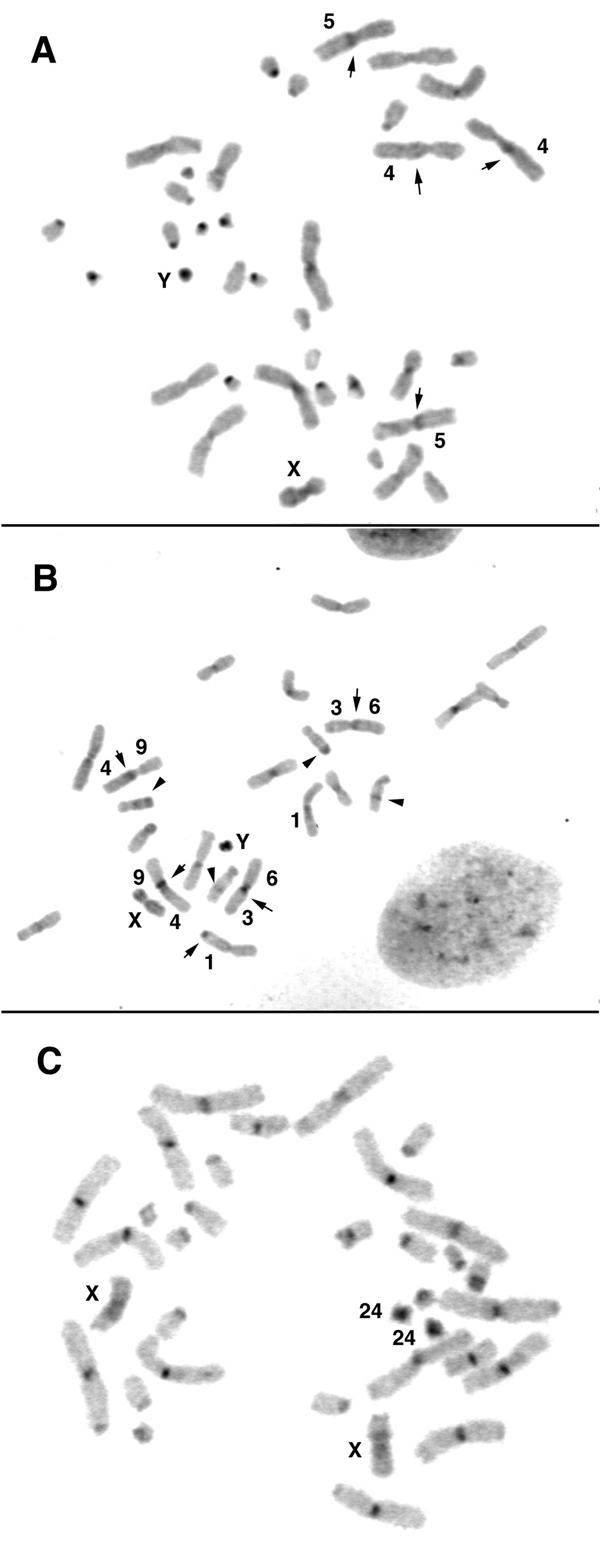
The composition of meta- and submetacentric autosomal pairs was analysed by G-band pattern comparison with the basic vespertilionid karyotype and with karyotypes of other Vespertilionini species. None of the four metacentric pairs of Myotis was conserved in N. happoldorum . Instead, the following arm combinations were found in pairs NHA1 to NHA8: 1/13, 2/7, 3/6, 4/9, 5/12, 8/10, 14/19, and 15/18. The composition of the three subtelocentric pairs was verified using MMY painting probes. NHA9 showed homology to three small Myotis chromosomes, MMY 23 in the short arm, MMY20 and MMY 22 in the long arm, separated by a small heterochromatic segment. NHA10 was homologous to the combined MMY24 and MMY25 painting probe in the short and MMY 11 in the long arm. FISH with tree shrew TBE30 probe confirmed homologous sequences to MMY 24 in the distal part of the NHA10 short arm. The smallest subtelocentric pair, NHA11, was shown to bear MMY21 homologous sequences in the short arm and the proximal part of the long arm, and MMY16/17 (TBE5) homologous sequences in the distal part of the long arm. The G-banding pattern of the X chromosome differed clearly from that of state II, which characterizes Vespertilionini and Pipistrellini species ( Fig. 7 View FIG ).
C-banding revealed a nearly completely heterochromatic Y chromosome and few small non-centromeric heterochromatic segments in addition to weak centromere staining. Only one homologue of NHA1 showed a terminally located heterochromatic band on the long arm. In NHA3 and NHA5, pericentromeric heterochromatin was present, but the homologs differed concerning the position, either in the short or the long arm. In NHA9, an interstitial C-positive band was present in the middle of the long arm, whereas a C-band positive segment was present at the long arm telomere in NHA11 on both homologs ( Fig. 6B View FIG ).
The NORs are positioned at the SC of the NHA8 long arm. Interestingly, NHA8, the fusion product of MMY18 and MMY15 homologs, showed a heteromorphism concerning the position of the centromere ( Fig. 9 View FIG ). One homologue conserved the ancestral condition concerning the position of NOR and centromere. In the second homologue, however, the centromere is located within the MMY18 homologous arm, resulting in a subtelocentric shape. As the G-banding pattern was preserved, a centromere repositioning event is assumed as mode of rearrangement. The chromosomes 11, 12, and 15 were present in vespertilionid state II. The baculum of this specimen is depicted in Fig. 10 View FIG .
No known copyright restrictions apply. See Agosti, D., Egloff, W., 2009. Taxonomic information exchange and copyright: the Plazi approach. BMC Research Notes 2009, 2:53 for further explanation.
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Tribe |
Vespertilionini |
|
Genus |