Exhippolysmata, , AND
|
publication ID |
https://doi.org/10.1111/zoj.12044 |
|
persistent identifier |
https://treatment.plazi.org/id/03C787FD-FFA6-9560-FC7E-46857C9B9E9B |
|
treatment provided by |
Marcus |
|
scientific name |
Exhippolysmata |
| status |
|
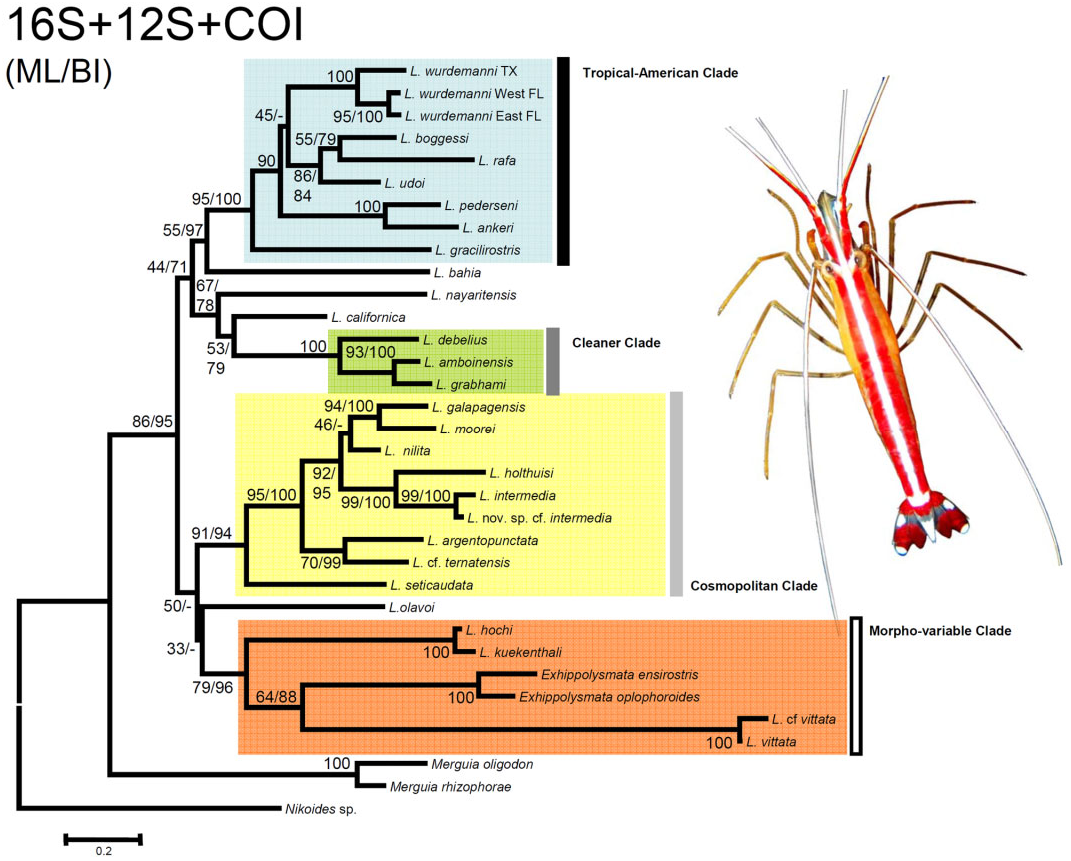
Phylogenetic trees using only reliable sequences (see above) obtained with ML and BI resulted in the same general topology ( Fig. 3 View Figure 3 ). In contrast to that reported by previous analyses, the phylogenetic analyses herein conducted did not support the genus Merguia as the sister group of a second natural group composed by shrimps from the genera Lysmata and Exhippolysmata . However, the monophyly of Merguia and of Lysmata + Exhippolysmata is well supported by a high posterior probability obtained from the BI analysis, and by bootstrap support from the ML analysis ( Fig. 3 View Figure 3 ). Also in contrast to that reported by previous analyses, the position of Lysmata olavoi as the most basally placed species within the monophyletic clade composed of species of Lysmata and Exhippolysmata is not supported by the ML and BI analyses. The species of Lysmata and Exhippolysmata can be subdivided into four monophyletic subclades.
The first monophyletic clade [well supported by ML (95) and BI (100) analyses], previously named ‘Neotropical’ or ‘Tropical American’, is composed of seven or eight species (if L. wurdemanni from the north Gulf of Mexico and from East Florida are considered two different species). Six or seven species inhabit the wider Caribbean and/or the Gulf of Mexico, and one species is from the Eastern Pacific ( Fig. 3 View Figure 3 ). Within this clade, the basal position of the Eastern Pacific L. gracilirostris is well supported by the BI (100) and ML (95) analyses. The only pair of species supported (both BI and ML values = 100) as sister taxa in this clade are Lysmata ankeri and L. pederseni .
The second monophyletic group [well supported by ML (100) and BI (100) analyses], previously named the ‘Cleaner’ clade, include three colourful and putatively fish-cleaning species: two ‘skunk’ or ‘lady scarlet’ shrimps, L. amboinensis and L. grabhami , that are well supported as sister taxa by ML (93) and BI (100) analyses within this group, and the red blood shrimp L. debelius , the basal position of which within the clade is also well supported ( Fig. 3 View Figure 3 ). Importantly, the topology of the trees indicate that two less colourful peppermint shrimps from the Eastern Pacific, Lysmata nayaritensis and L. californica , are related, and have a basal position within this clade; however, ML bootstrap and BI support values for the position of these two latter species is only moderate ( Fig. 3 View Figure 3 ).
The third clade, named the ‘Cosmopolitan’ clade [well supported by ML (91) and BI (94) analyses], is composed of nine species: two from the Mediterranean ( Lysmata nilita and L. seticaudata ), three from the western Atlantic ( Lysmata moorei , Lysmata intermedia , and Lysmata cf. intermedia sp. nov.), three from the tropical eastern Pacific ( Lysmata galapagensis , Lysmata argentopunctata , and Lysmata holthuisi ), and one from the Indo-Pacific ( Lysmata cf. ternatensis). Two pairs of species ( L. intermedia and L. cf. intermedia sp. nov.; L. galapagensis and L. moorei ) are well supported as sister taxa by ML and BI analyses ( Fig. 3 View Figure 3 ).
The fourth clade, named ‘Morphovariable’, was retrieved from both ML and BI phylogenetic analyses; however, high support for the monophyly of this clade was obtained only from the BI analysis [the ML bootstrap value was moderate (79)]. This clade is composed of three pairs of well-supported sister taxa: the Caribbean Lysmata hochi and the Indo-Pacific Lysmata kuekenthali , the Indo-Pacific Lysmata vittata , and the Caribbean Lysmata cf. rauli, and two shrimps from the genus Exhippolysmata ( Exhippolysmata oplophoroides from the western Atlantic and Exhippolysmata ensirostris from the Indo-Pacific) ( Fig. 3 View Figure 3 ).
Phylogenetic trees obtained with ML and BI but conducted using a single mitochondrial marker resulted in somewhat similar general topologies ( Fig. 4). Most of these single-gene analyses retrieved the monophyletic clades observed in the multigene tree described above. Nonetheless, in some cases, the separation of the species on the different monophylectic clades was only partial. For instance, in both the ML and the BI trees based on the 12S gene fragment, L. pederseni and L. ankeri were well supported as sister species, but were not included within the Neotropical clade retrieved during the total-evidence analysis ( Fig. 4).
PHYLOGENETIC INFORMATIVENESS OF
No known copyright restrictions apply. See Agosti, D., Egloff, W., 2009. Taxonomic information exchange and copyright: the Plazi approach. BMC Research Notes 2009, 2:53 for further explanation.