Apatania zonella
|
publication ID |
https://doi.org/10.1093/isd/ixab013 |
|
persistent identifier |
https://treatment.plazi.org/id/03C887EB-FFD8-5218-FC83-1219153B83CA |
|
treatment provided by |
Felipe |
|
scientific name |
Apatania zonella |
| status |
|
Apatania zonella View in CoL Group
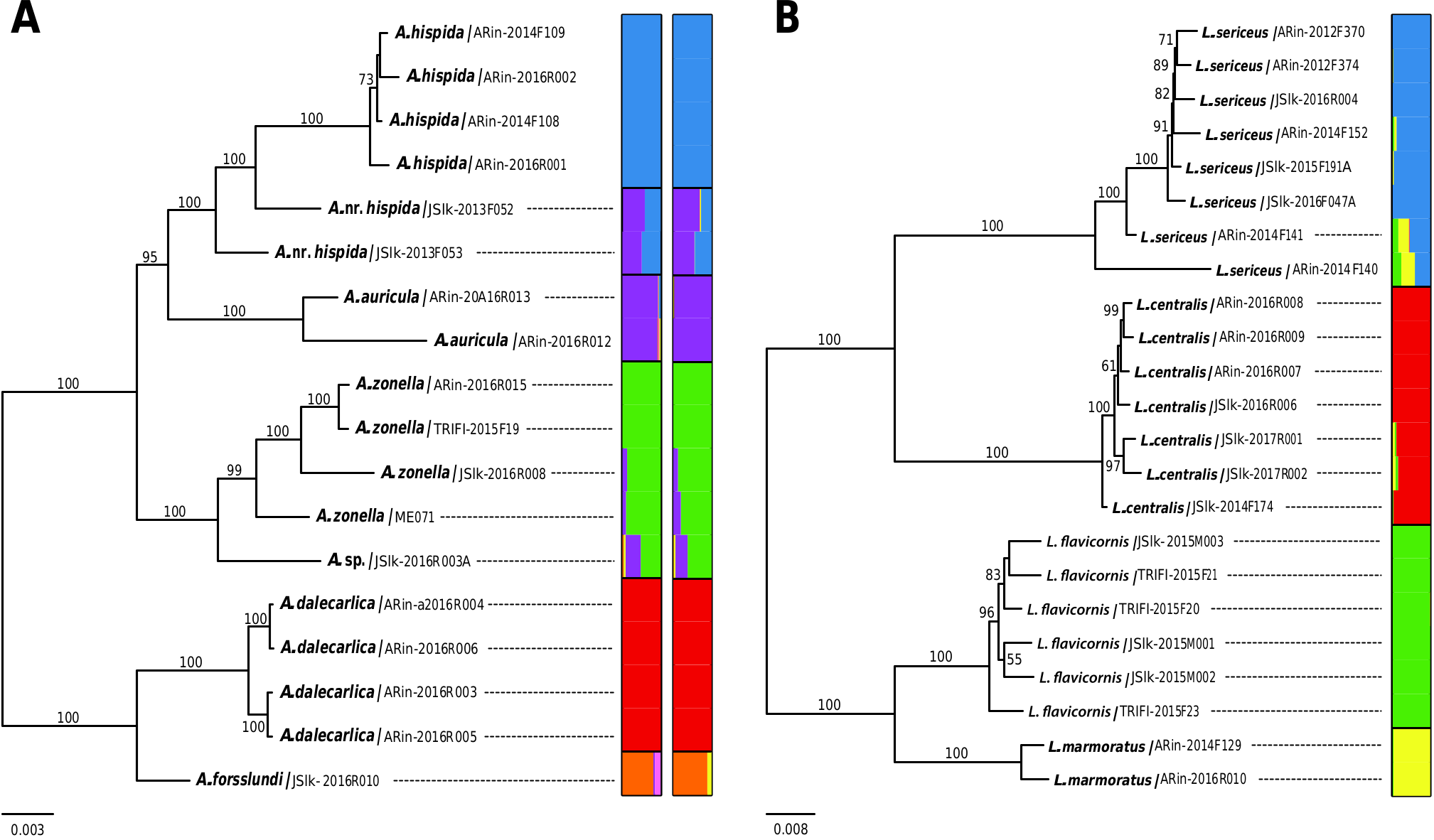
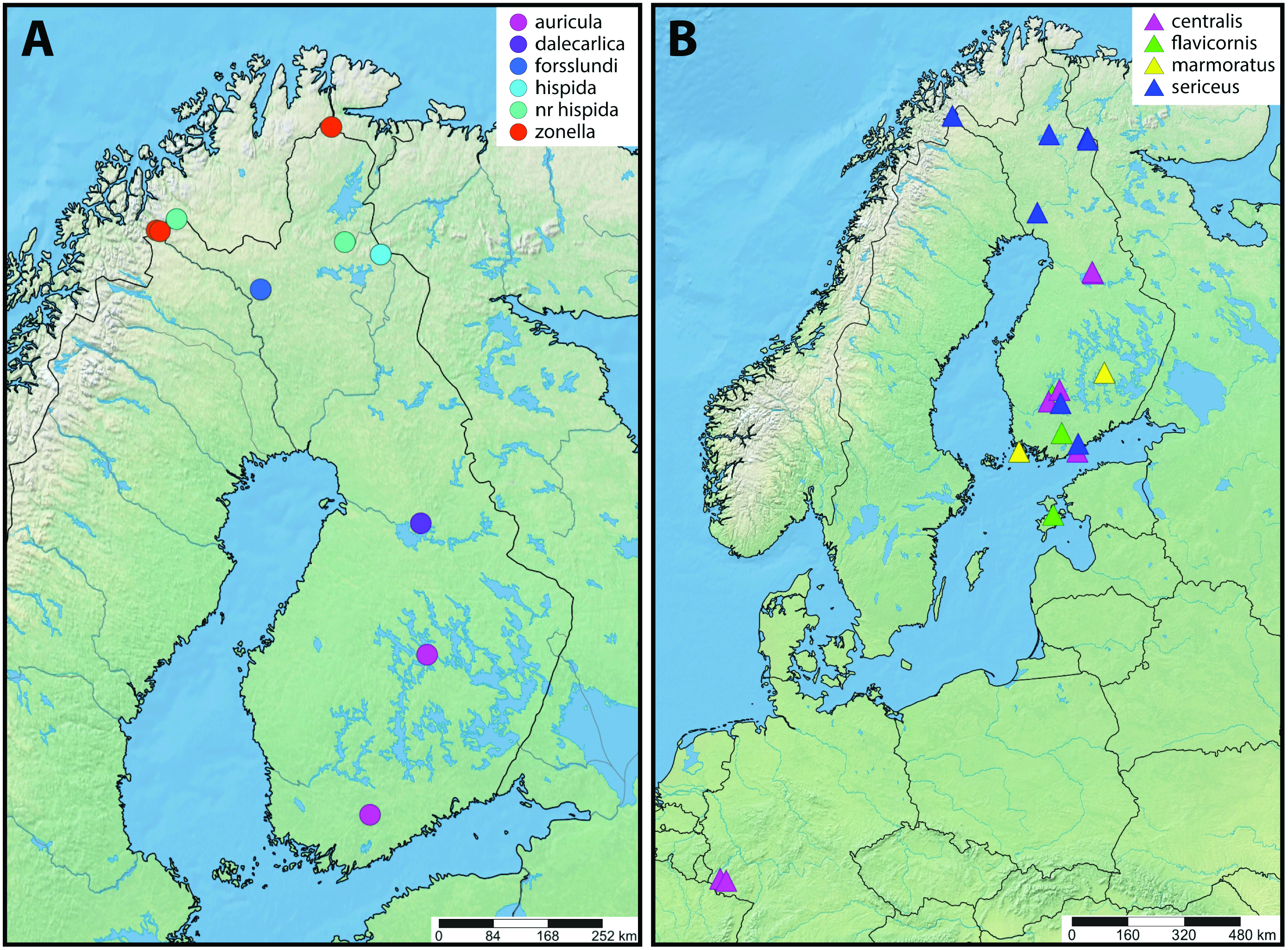
Species recognition using the morphological characters was in line with the key provided by Salokannel et al. (2010), except for the specimen JSlk-2016R003A. Apatania zonella group specimens included in the analysis represent the five valid Finnish species and A. nr. hispida . The specimen JSlk-2016R003A has a ventrally bluntrounded supragenital plate similar to A. zonella , but the upper part of the last segment is protruded, making it laterally similar to A. nr. hispida . Such combination of characters is not clearly associated with any valid taxon, so the sample was named as Apatania sp.
Sequence variation of mtCOI within species was close to zero (0–0.15%) except in A. zonella with 1.16% variation. A. hispida and A. nr. hispida samples have identical DNA barcode. Apatania sp. (JSlk-2016R003A) has the closest match (0.15%) to the specimens of A. forsslundi . Otherwise, the inter-species variation between the valid species ranges from 1.05 to 2.31% ( Table 2).
We generated a genome-wide SNP dataset from 18 individuals of the A. zonella group using ddRAD sequencing. We obtained 3.2 million reads per individual on average, of which 3 million reads per individual (93.3%) were retrieved after quality filtering step
NA, not applicable, single specimen.
(Supp Table 1 [online only]). After clustering at 85% sequence similarity, 26,790 putative orthologous loci shared across more than four samples were retained, for a total length of 2,158,568 bp ( Table 3). These data included 59,486 SNPs, of which 38,366 are parsimony informative. The proportion of missing data was 71.4% across all loci.
At the genomic level (ddRAD data), within and between species divergences partly overlapped in A. zonella group. Within species divergences ranged from 0.12 ( A. hispida ) to 0.66% ( A. nr. hispida ), whereas the mean interspecific distances varied from 0.64 (between A. nr. hispida and A. forsslundi ) to 2.91% (between A. hispida and A. dalecarlica ) ( Table 2).
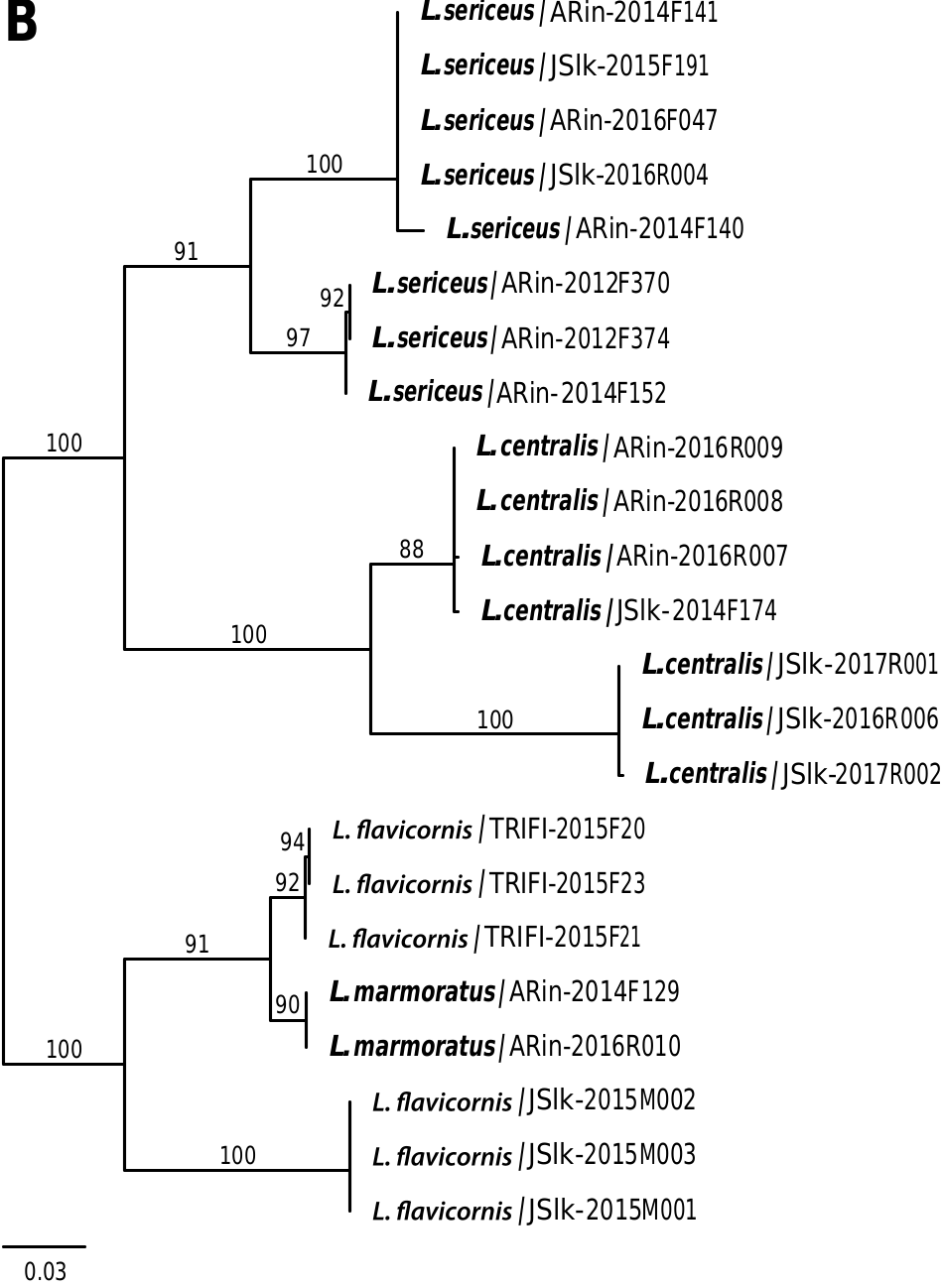
In the ML tree based on mtCOI sequences, most species in the A. zonella group did not clearly separate as distinct clades, except for A. auricula ( Fig. 2A View Fig ). Apatania nr. hispida completely intermixed with A. hispida , while A. zonella diverged into two lineages and was found closely related to A. dalecarlica . In the ddRAD ML tree, the lineages that correspond to A. hispida , A. auricula , and A. dalecarlica were strongly supported as monophyletic (BS 100%) ( Fig. 3A View Fig ). Apatania nr. hispida appeared paraphyletic with respect to its putative sister taxon A. hispida . The unidentified sample, Apatania sp. , was closely related to A. zonella . The species tree estimation analyses recovered a rather similar topology to the ML tree, but A. auricula was found sister to A. zonella and Apatania sp. (Supp Fig. 1 View Fig [online only]).
The population clustering analyses revealed substantial heterogeneity in proportions of admixed ancestry between the species. The best supported model ( K = 6) clustered populations into six major clades ( Fig. 3A View Fig ; Supp Fig. 2 View Fig [online only]). The STRUCTURE analysis revealed that much of common ancestry of Apatania nr. hispida is shared through apparent admixture with the closely related See details for all data sets in Supp Table S1 (online only). 0.006
A. hispida and A. auricula ( Fig. 3A View Fig ). However, D-statistics estimated insignificant levels of gene flow between A. hispida and A. auricula , but significant only between A. hispida and A. nr. hispida (Supp Table 3 [online only]). Apatania sp. also shared significant common ancestry with A. zonella and A. auricula , and did not form a distinct cluster even when the number of assumed genetic clusters was increased to 7.
No known copyright restrictions apply. See Agosti, D., Egloff, W., 2009. Taxonomic information exchange and copyright: the Plazi approach. BMC Research Notes 2009, 2:53 for further explanation.