Tingidae, Laporte, 1833
|
publication ID |
https://doi.org/10.11646/zootaxa.4722.5.3 |
|
publication LSID |
lsid:zoobank.org:pub:0183A47A-AA1E-4AAF-8802-54CB9CCDE58C |
|
persistent identifier |
https://treatment.plazi.org/id/03CCC759-F00C-1C07-FF74-F97324CEFF50 |
|
treatment provided by |
Plazi |
|
scientific name |
Tingidae |
| status |
|
Phylogeny of Tingidae View in CoL View at ENA
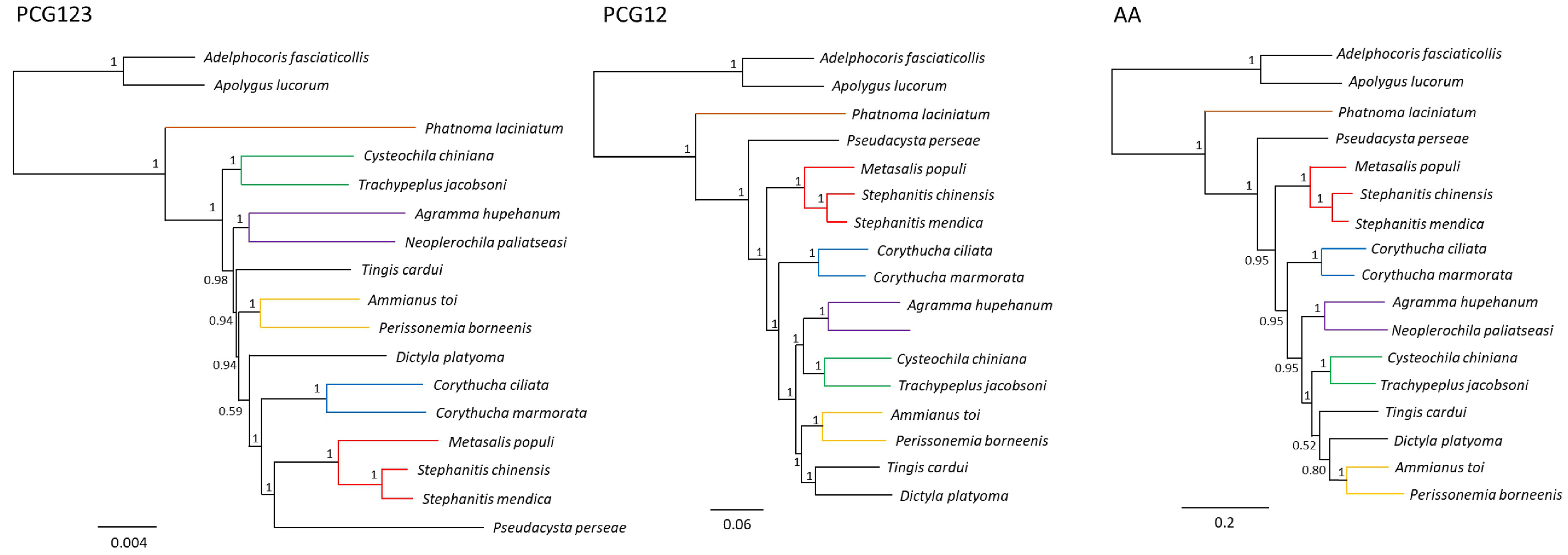
Phylogenetic relationships among Tingidae were inferred including all the mitogenomes available, except Eteoneus sigillatus Drake & Poor 1936 ( KU896784 View Materials , unverified sequence). This dataset represents the highest mitogenomic coverage of Tingidae so far, although it represents a mere 0.6% of the family (14 species in 12 genera), all belonging to the subfamily Tinginae . We tested three datasets: PCG123 with each codon position partitioned, PCG12 (only the 1 st and the 2 nd codon positions, and AA (amino acid sequence. PCG12 and AA were tested as this approach was shown to decrease the impact of the high compositional heterogeneity typical of Heteroptera mitogenomes ( Liu et al., 2018; Yang et al., 2018).
Statistical support was higher in the PCG12 tree (BPP = 1, all nodes) than in the AA or the PCG123 trees, where several nodes had a BPP lower than 1 ( Figure 7 View FIGURE 7 ). The PCG12 and AA trees showed a very similar topology, differing only in the positions of Tingis cardui and Dictyla platyoma (Fieber 1861) , which also had the lowest statistical support of all nodes in the AA tree. Although the order of some deeper nodes differed between the PCG123, PCG12 and AA trees, all topologies showed Phatnoma laciniatum (Phatnomini) in the basal position to all other species ( Tingini ), as expected, and Stephanitis and Corythucha , the only genera represented by more than one species, as monophyletic. These two features were also recovered in previous phylogenies ( Guilbert, Damgaard and D’Haese, 2014; Lin et al., 2017). Metasalis populi was not recovered as sister to Tingis cardui , in disagreement with their putative close relationship, as M. populi was previously described as a Tingis species. Instead, M. populi was sister to the Stephanitis clade, and Cysteochila chiniana Drake 1954 was sister to Trachypeplus jacobsoni Horváth 1926 , as previously found ( Yang et al., 2018). Ammianus toi (Drake 1938) and Perissonemia borneenis (Distant 1909) were also sister taxa in the all trees. Neoplerochila paliatseasi was most closely related to Agramma hupehanum (BPP = 1, in all trees), in accordance to the high similarity between the COI sequences of the two species, which was the criteria we used for selecting A. hupehanum as the reference mitogenome for the mapping and assembly of N. paliatseasi . Agramma Stephens 1829 is considered to be basal to Tingini in traditional systematics works (see Peìricart, 1983), and A. hupehanum was also recovered as basal in phylogenies based on shorter gene regions and morphological characters ( Guilbert, Damgaard and D’Haese, 2014). However, this was not the case in our study, similarly to Liu et al. 2018, and the basal position in in the PCG12 and AA trees was occupied by Pseudacysta perseae (as in Kocher et al. 2015). Several studies have presented reconstructions of the phylogeny of true bugs, including Tingidae , based on mitochondrial genomes ( Yang, Yu and Du, 2013; Kocher et al., 2015; H. Li et al., 2017; Lin et al., 2017; P. W. Li et al., 2017; Liu et al., 2018; Yang et al., 2018). However, the relationships within Tingidae could not be discussed as too few terminals for the family were included. Mitogenomic phylogenies have recovered different topologies compared to phylogenies based on shorter mitochondrial regions combined with morphological characters ( Guilbert, Damgaard and D’Haese, 2014), and the results have been contradictory. This could be explained by the large number of characters (up to 15,000 bp) generally analyzed in mitogenomic phylogenies on a few terminal taxa, in contrast with the analysis of a much higher number of taxa using shorter sequences (up to 5,000 bp) ( Guilbert, Damgaard and D’Haese, 2014). Additionally, the inconsistent recovery of the phylogeny of Tingidae could also be explained by the high sequence heterogeneity and the highest evolutionary rate characteristic of this family among Cimicomorpha, and the highest family-level nonsynonymous substitutions per non-synonymous site (Ka) among true bugs ( Liu et al., 2018). In line with previous studies, our phylogenetic reconstruction confirmed that the order of the deeper nodes varies depending on the type of analyses performed. In the future, it may be necessary to include nuclear DNA, as mitochondrial data does not produce consistent results.
No known copyright restrictions apply. See Agosti, D., Egloff, W., 2009. Taxonomic information exchange and copyright: the Plazi approach. BMC Research Notes 2009, 2:53 for further explanation.
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |