Eulaemus, Girard, 1857
|
publication ID |
https://doi.org/10.1111/zoj.12231 |
|
persistent identifier |
https://treatment.plazi.org/id/03E02B2A-FFCA-4A39-D6AA-C044FD3FFC45 |
|
treatment provided by |
Felipe |
|
scientific name |
Eulaemus |
| status |
|
EULAEMUS PHYLOGENY
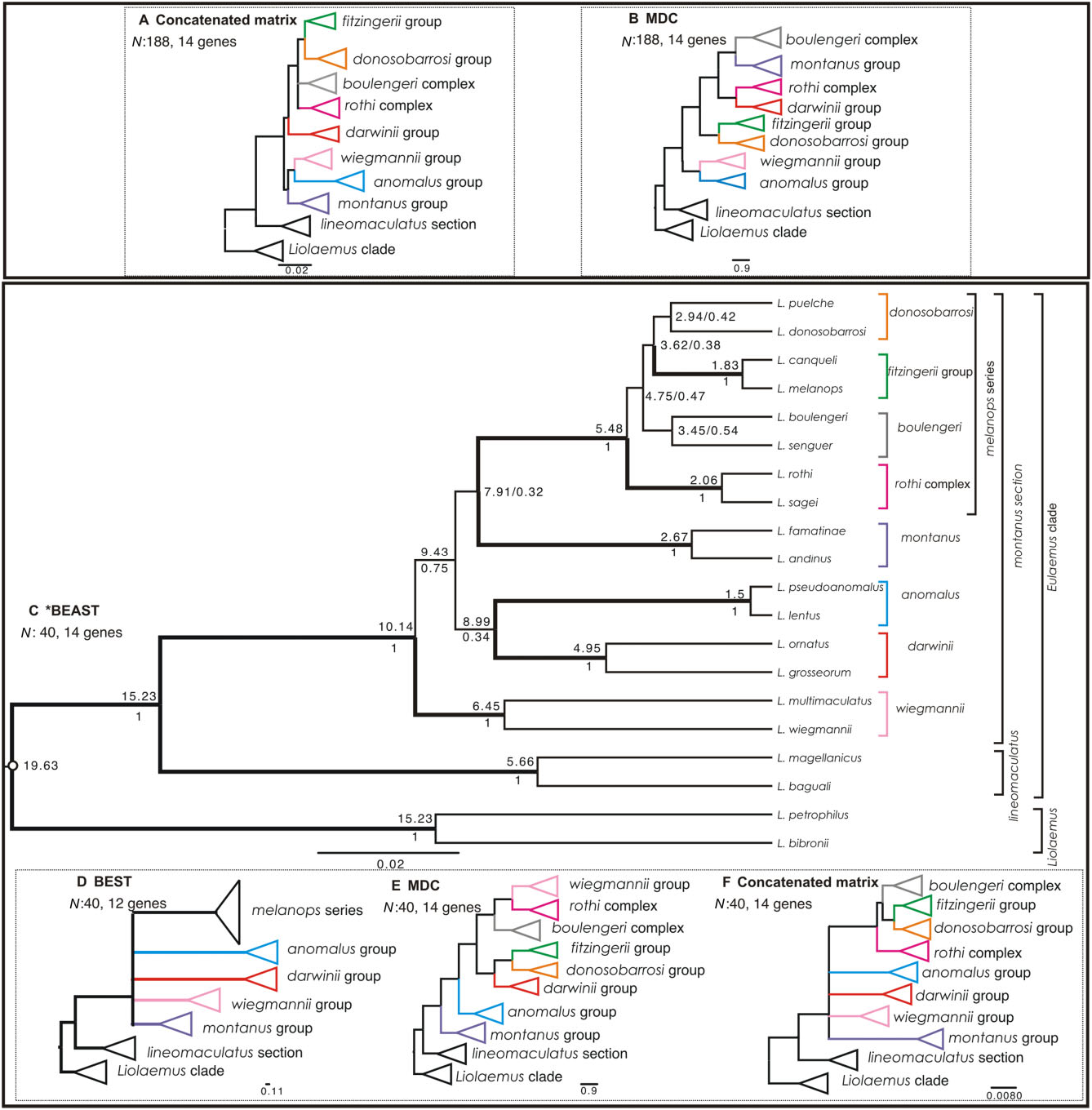
Using a reduced matrix of 40 taxa and 14 loci, the main clades were recovered as monophyletic using every method. However, with the exception of the lineomaculatus section, which was recovered as sister clade of the montanus section under every method, we found strong discordances amongst phylogenetic estimates of the main clades.
1. Our *BEAST analysis ( Fig. 3C View Figure 3 ) recovered all of the main groups as monophyletic and with high statistical support (PP = 1), except for the boulengeri complex and donosobarrosi group, which had no support. We again recovered the L. lineomaculatus section (PP = 1) as the sister clade to the L. montanus section (PP = 1), with strong support (PP = 1). Within the montanus section, we recovered the melanops series ( rothi complex + boulengeri complex + fitzingerii group + donosobarrosi group) as a well-supported clade, but relationships amongst these are not resolved (PP <0.47), and monophyly is strongly supported only for the fitzingerii group and the rothi complex. The wiegmannii , darwinii , anomalus , and montanus groups are strongly supported as monophyletic, but relationships between these and the melanops series are not resolved (PP <0.75).
2. BEST ( Fig. 3D View Figure 3 ). The species tree based on 12 nuclear loci obtained with BEST is highly concordant with the one obtained using *BEAST with 14 loci (including the two mitochondrial loci). We recovered the same pattern of strongly supported main clades and the lineomaculatus section as the sister clade to all others, which were unresolved.
3. MDC ( Fig. 3E View Figure 3 ). The lineomaculatus section is consistently recovered as the sister clade to all others in the Eulaemus clade. The montanus group is recovered in a basal position within the montanus section, and the anomalus group was recovered as sister of [(( darwinii group + ( donosobarrosi group + fitzingerii group)) + (( boulengeri complex + ( rothi complex + wiegmannii group))]. This topology differs from the tree recovered by the MDC analysis of the complete data set ( Fig. 3B View Figure 3 ).
4. Concatenated analysis ( Fig. 3F View Figure 3 ). These results are concordant with the *BEAST and BEST trees ( Fig. 3C, D View Figure 3 ), but here we recovered full resolution and higher support values for clades within the melanops series (PP = 1 in all cases). We obtained similar no- to low-support values for relationships amongst the wiegmannii , darwinii , anomalus , and montanus groups plus the melanops series.
No known copyright restrictions apply. See Agosti, D., Egloff, W., 2009. Taxonomic information exchange and copyright: the Plazi approach. BMC Research Notes 2009, 2:53 for further explanation.