Litinium gludi, Leduc & Zhao, 2021
|
publication ID |
https://doi.org/10.5852/ejt.2021.748.1347 |
|
publication LSID |
lsid:zoobank.org:pub:E814BAB1-3B3D-4458-9F94-E1EF89191881 |
|
DOI |
https://doi.org/10.5281/zenodo.4746299 |
|
persistent identifier |
https://treatment.plazi.org/id/FA79DFEF-A389-48AA-AF75-03984D69F202 |
|
taxon LSID |
lsid:zoobank.org:act:FA79DFEF-A389-48AA-AF75-03984D69F202 |
|
treatment provided by |
Plazi |
|
scientific name |
Litinium gludi |
| status |
sp. nov. |
Litinium gludi sp. nov.
urn:lsid:zoobank.org:act:
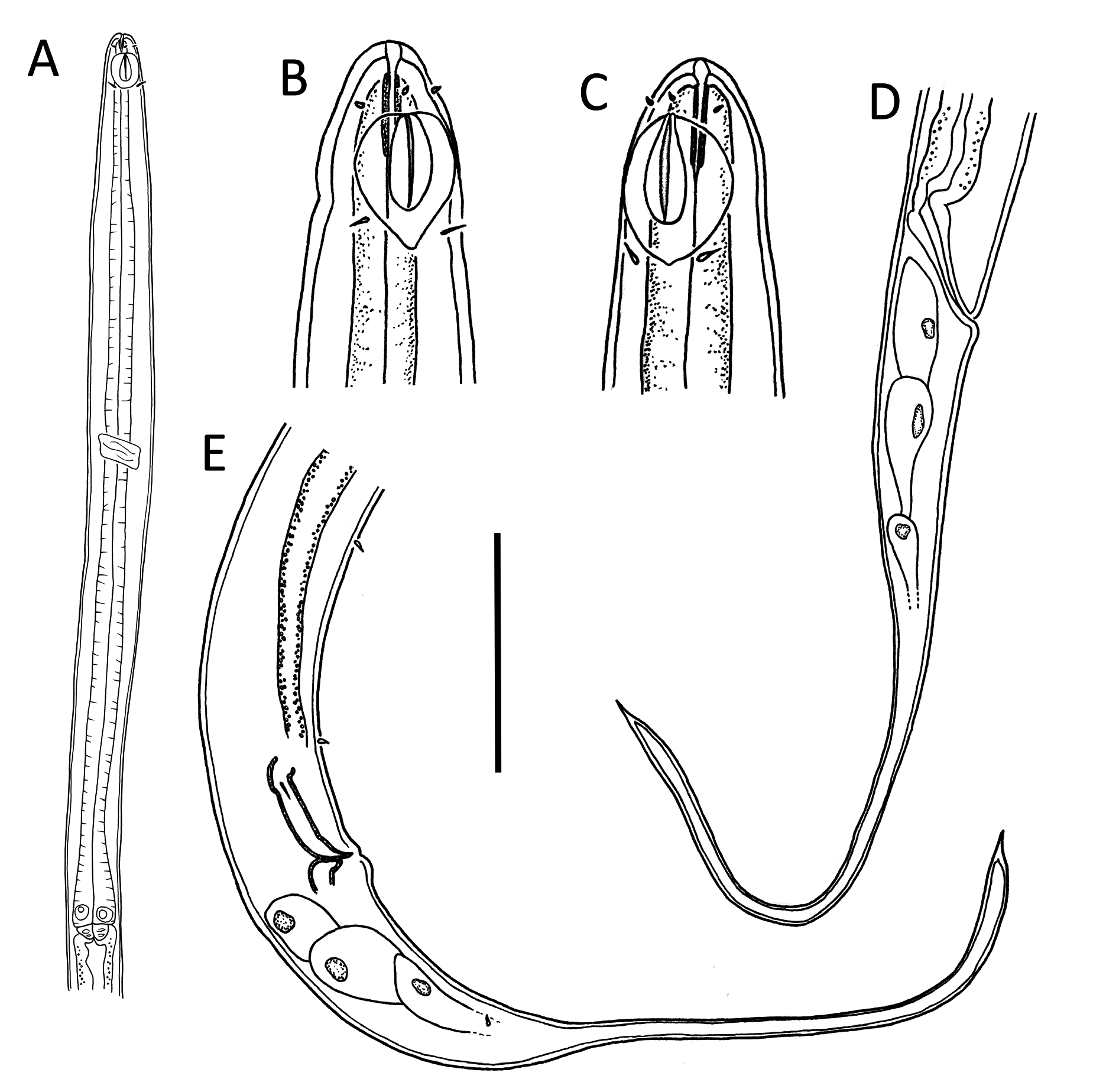
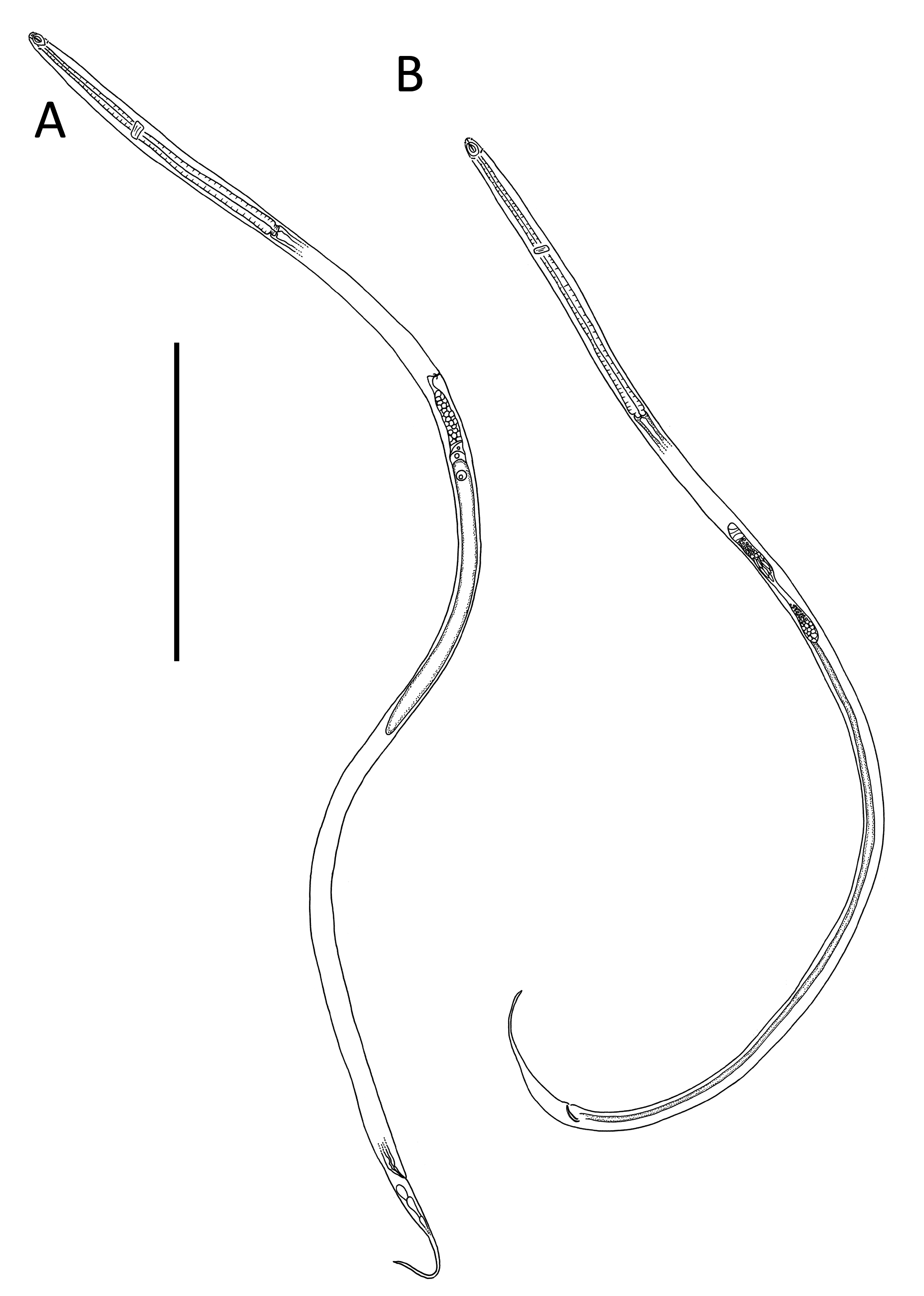
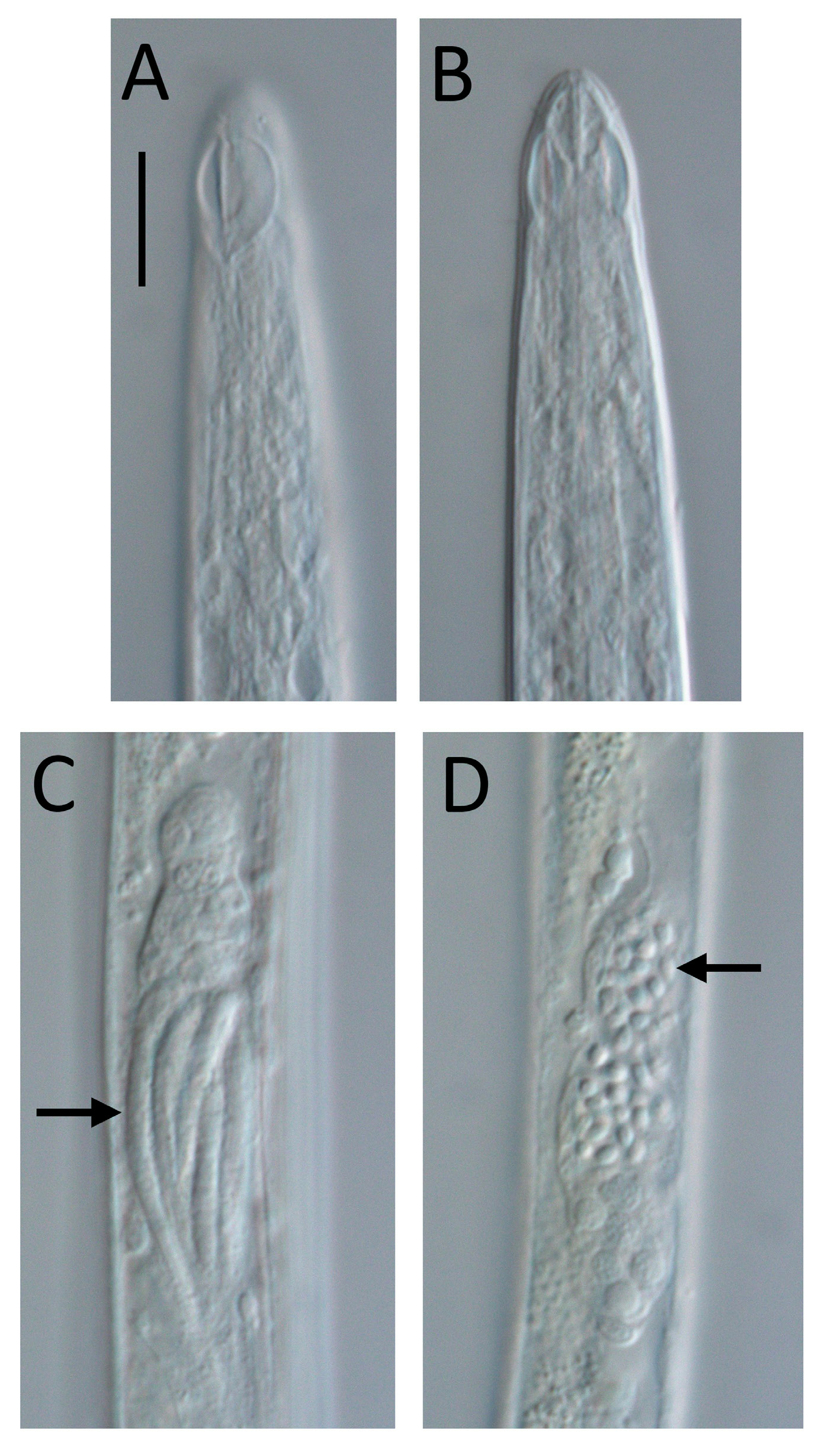
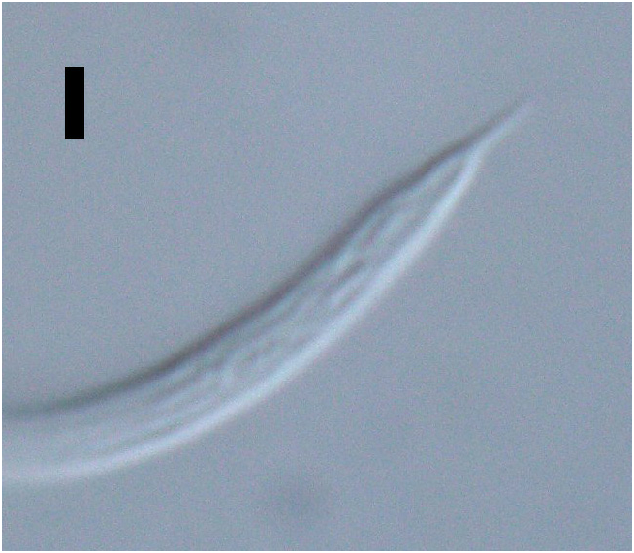
Figs 1–4 View Fig View Fig View Fig View Fig , Table 1 View Table 1
Diagnosis
Litinium gludi sp. nov. is characterised by a slender body (a = 58–63), a body length of 895–1066 µm, papilliform labial sensilla, short cephalic sensilla 1.5–2.0 µm or ~0.2 cbd long, a large heart- or leafshaped amphideal fovea 67–88% cbd wide, a smaller amphideal aperture with two central longitudinal ridges, males with dimorphic sperm, spicules 1.1–1.4 times cloacal body diameter, a short and simple gubernaculum, and two precloacal setae.
Differential diagnosis
The new species resembles L. aceratus and L. longicaudatus , the only other species of the genus with conico-cylindrical tails, and is particularly similar to L. longicaudataus in having papillose (< 2 µm) labial sensilla. Litinium gludi sp. nov. can be differentiated from L. longicaudatus by the shorter body (895–1066 vs 2180–2196 µm in L. longicaudatus ), lower ‘a’ ratio (58–63 vs 75–86), and heart- or leafshaped amphideal fovea (vs pocket-shaped).
Etymology
The species is named after Professor Ronnie Glud, leader of the HADES-ERC trench project and covoyage leader.
Material examined
Holotype PACIFIC OCEAN • ♂; Kermadec Trench , voyage TAN1711, station 25, site 3; 30.3815° S, 176.6417° W; 9540 m water depth, 0–2 cm sediment depth; 3 Dec. 2017; NIWA 139257 View Materials . GoogleMaps
Paratypes PACIFIC OCEAN • 2 ♂♂, 1 ♀; Kermadec Trench , voyage TAN1711, station 25, site 3; 30.3815° S, 176.6417° W; 9540 m water depth, 0–2 cm sediment depth; 3 Dec. 2017; NIWA 139258 View Materials GoogleMaps .
Description
Males
Body slender, cylindrical, widest at level of nerve ring, tapering slightly towards anterior extremity. Cuticle smooth, without ornamentation; somatic setae absent, except for sparse sublateral setae on tail. Cephalic region demarcated by slight constriction near posterior edge of amphideal fovea. Inner labial sensilla not observed; six outer labial papillae, ~ 0.5 µm long, located slightly anterior to amphideal fovea and four short cephalic sensilla, 1.5–2.0 µm or ~0.2 cbd long, situated far posteriorly at level of posterior edge of amphideal fovea. Amphideal fovea large, heart- or drop-shaped, with lightly cuticularized outline; amphideal aperture smaller, oval- or drop shaped, with two lightly cuticularized longitudinal central ridges spanning long axis of amphideal fovea. Buccal cavity narrow, cylindrical, with cuticularized portion 3–5 µm deep. Pharynx muscular, cylindrical, surrounding posterior portion of buccal cavity, widening towards posterior extremity but not forming true bulb. Nerve ring located slightly anterior to middle of pharynx. Secretory-excretory system not observed. Cardia 2–4 µm long, partially embedded in intestine.
Reproductive system with two opposed and outstretched testes; anterior testis located to the left or right of intestine, posterior testis located on opposite side or on same side. Mature sperm in anterior testis large, elongated, 19–21 × 2–3 µm; mature sperm in posterior testis smaller, globular, 1.5 × 1.5–2.0 µm. Spicules short, 1.1–1.4 times cloacal body diameter, curved distally, with ventrally bent capitulum. Gubernaculum small, simple, block-shaped, without lateral pieces, with poorly developed apophyses. Two precloacal papillae, 1.3–1.4 µm long; posterior-most papilla located 11–14 µm anterior to cloaca, second precloacal papilla located 19–22 µm from posterior-most precloacal papilla. Ejaculatory glands not observed. Tail conico-cylindrical, with pointed tip, caudal capsule absent; three caudal glands present posterior to cloaca; spinneret not observed.
Female
Similar to males but without any somatic setae on tail. Reproductive system with single posterior genital branch and reflexed ovary. Oocyte up to 160 × 10 µm. Vulva located far anteriorly at about ⅓ of body length from anterior extremity. Vagina perpendicular to body surface; vaginal glands not observed.
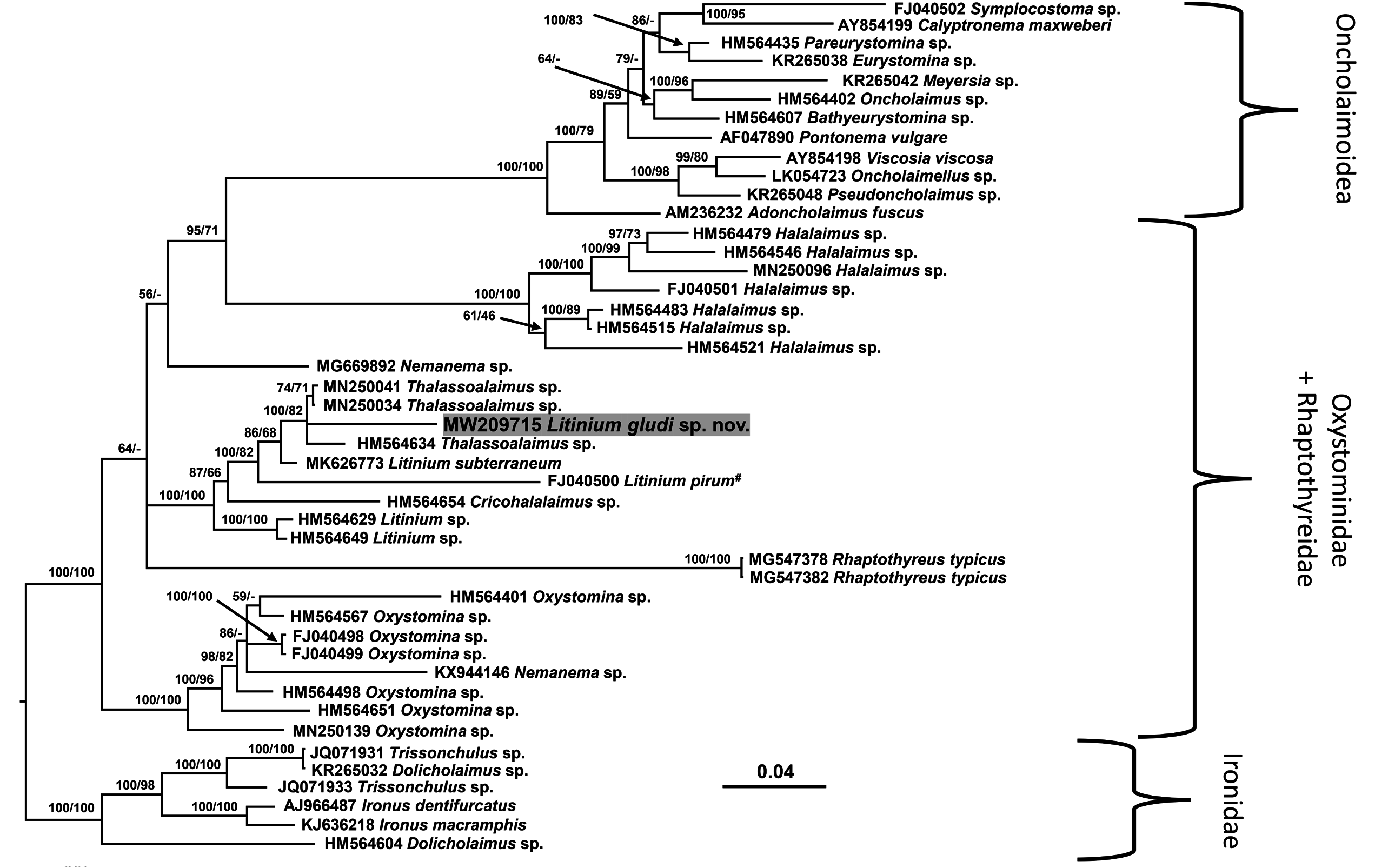
Molecular phylogenetic relationships
Near full-length SSU (1516 bp) and D2-D3 of LSU sequences (756 bp) were obtained for Litinium gludi sp. nov. The Oxystominidae did not form a monophyletic clade in the consensus SSU tree due to the placement of Rhaptothyerus typicus Hope & Murphy, 1969 and oncholaimoid sequences among the oxystominid sequences ( Fig. 5 View Fig ). Neither the Halalaiminae (represented by Halalaimus and Oxystomina ) nor the Oxystomininae (all other oxystominid genera) were recovered as monophyletic. Litinium gludi sp. nov. was placed in a well-supported clade (100% posterior probability and 82% bootstrap support) with sequences of Thalassoalaimus . The similarity between SSU sequences of Litinium gludi sp. nov. and Thalassoalaimus sp. was 94–95% with a difference of 69 in 1508 nucleotides with 4 gaps ( MN250034 View Materials ), 83 in 1504 nucleotides with 1 gap ( HM564634 View Materials ) and 43 in 798 nucleotides with 1 gap ( MN250041 View Materials ). The Litinium gludi sp. nov. + Thalassoalaimus clade was part of a wider, strongly supported oxystominid clade including all sequences of Litinium and Cricohalalaimus (100% posterior probability and bootstrap support), although neither Litinium nor Thalassoalaimus were monophyletic. The similarity between SSU sequences of Litinium gludi sp. nov. and Litinium sp. was 87–94% with a difference of 84 in 1513 nucleotides with 5 gaps ( FJ040500 View Materials ), 116 in 1516 nucleotides with 3 gaps ( HM564629 View Materials ), 113 in 1514 nucleotides with 2 gaps ( HM564649 View Materials ), and 46 in 786 nucleotides with 1 gap ( MK626773 View Materials ). Sequences of Halalaimus formed a strongly supported (100% posterior probability and bootstrap support) monophyletic sister group to the Oncholaimoidea Filipjev, 1916, the latter also forming a strongly supported monophyletic group (100% posterior probability and bootstrap support). The sequences of Oxystomina formed a strongly supported group (100% posterior probability and bootstrap support), however this genus was not monophyletic due to the inclusion of a sequence of Nemanema Cobb, 1920 in the clade. The placement of another sequence of Nemanema as a sister group to the Halalaimu s + Oncholaimoidea clade was poorly supported (56% posterior probability and <50% bootstrap support).
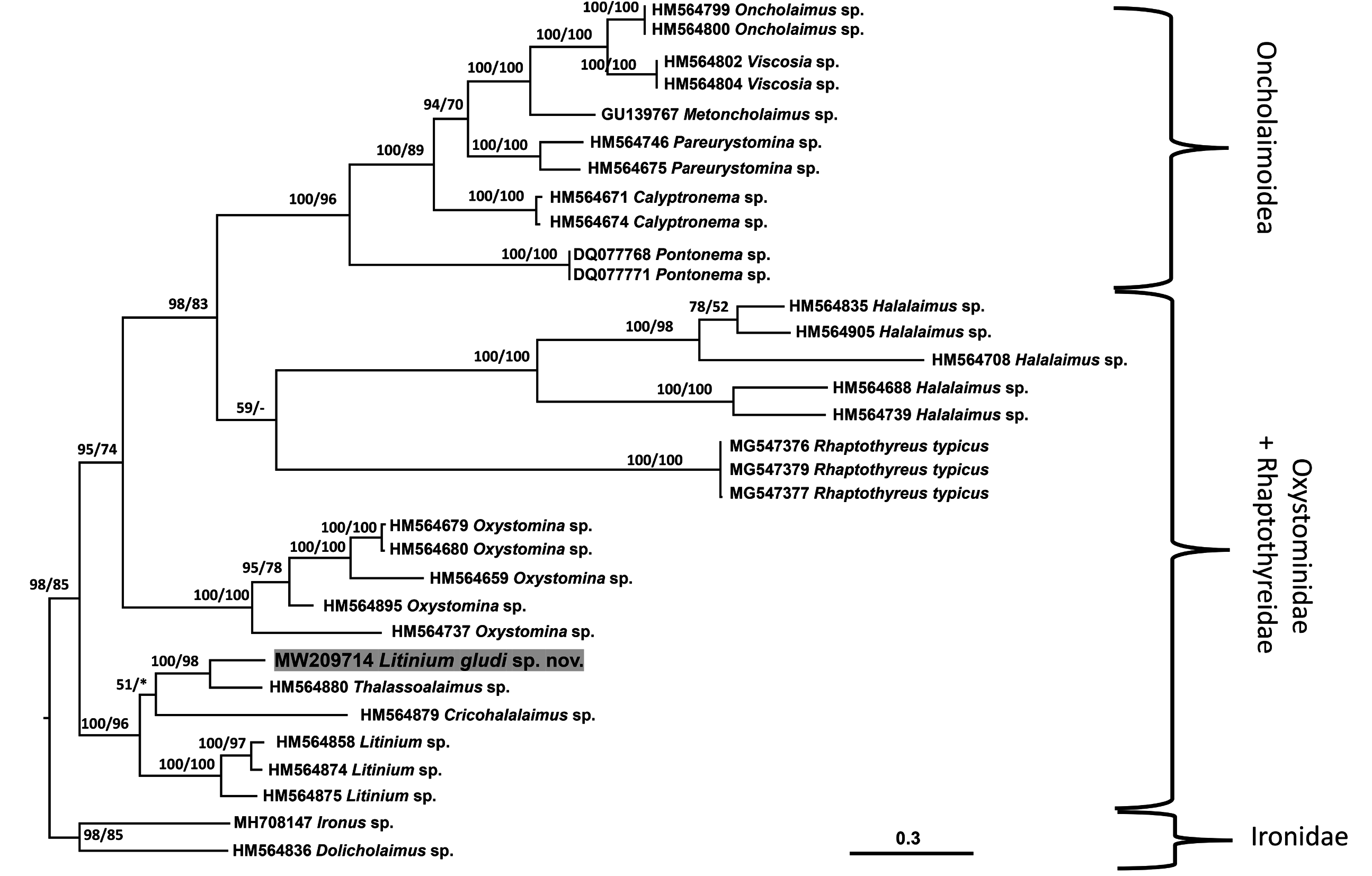
The topology of the D2-D3 of LSU consensus tree was similar to that of the SSU tree, with the Oxystominidae again not recovered as monophyletic ( Fig. 6 View Fig ). Sequences of Halalaimus and Oxystomina each formed strongly supported monophyletic clades (100% posterior probability and bootstrap support). As in the SSU tree, Litinium, Cricohalalaimus, and Thalassohalaimus formed a strongly supported clade (100% posterior probability and bootstrap support), with Litinium gludi sp. nov. most closely related to Thalassoalaimus (100% posterior probability and 98% bootstrap support). The similarity between LSU sequences of Litinium gludi sp. nov. and Thalassoalaimus sp. was 92% with a difference of 118 in 664 nucleotides with 7 gaps ( HM564880 View Materials ); the similarity between LSU sequences of Litinium gludi sp. nov. and Litinium sp. was 71–72% with a difference of 188 in 671 nucleotides with 13 gaps ( HM564858 View Materials ), 184 in 671 nucleotides with 12 gaps ( HM564874 View Materials ) and 183 in 671 nucleotides with 13 gaps ( HM564875 View Materials ).
No known copyright restrictions apply. See Agosti, D., Egloff, W., 2009. Taxonomic information exchange and copyright: the Plazi approach. BMC Research Notes 2009, 2:53 for further explanation.
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
SubOrder |
Ironina |
|
SuperFamily |
Ironoidea |
|
Family |
|
|
SubFamily |
Oxystomininae |
|
Genus |