Spartina, Schreb.
|
publication ID |
https://doi.org/10.1016/j.phytochem.2020.112312 |
|
DOI |
https://doi.org/10.5281/zenodo.8302676 |
|
persistent identifier |
https://treatment.plazi.org/id/03E487FB-FFC6-FF9E-4175-FA58FB46FDD1 |
|
treatment provided by |
Felipe |
|
scientific name |
Spartina |
| status |
|
2.1. Determination of the foliage phenolic fingerprint of Spartina View in CoL species by HPLC-DAD and LC/MS
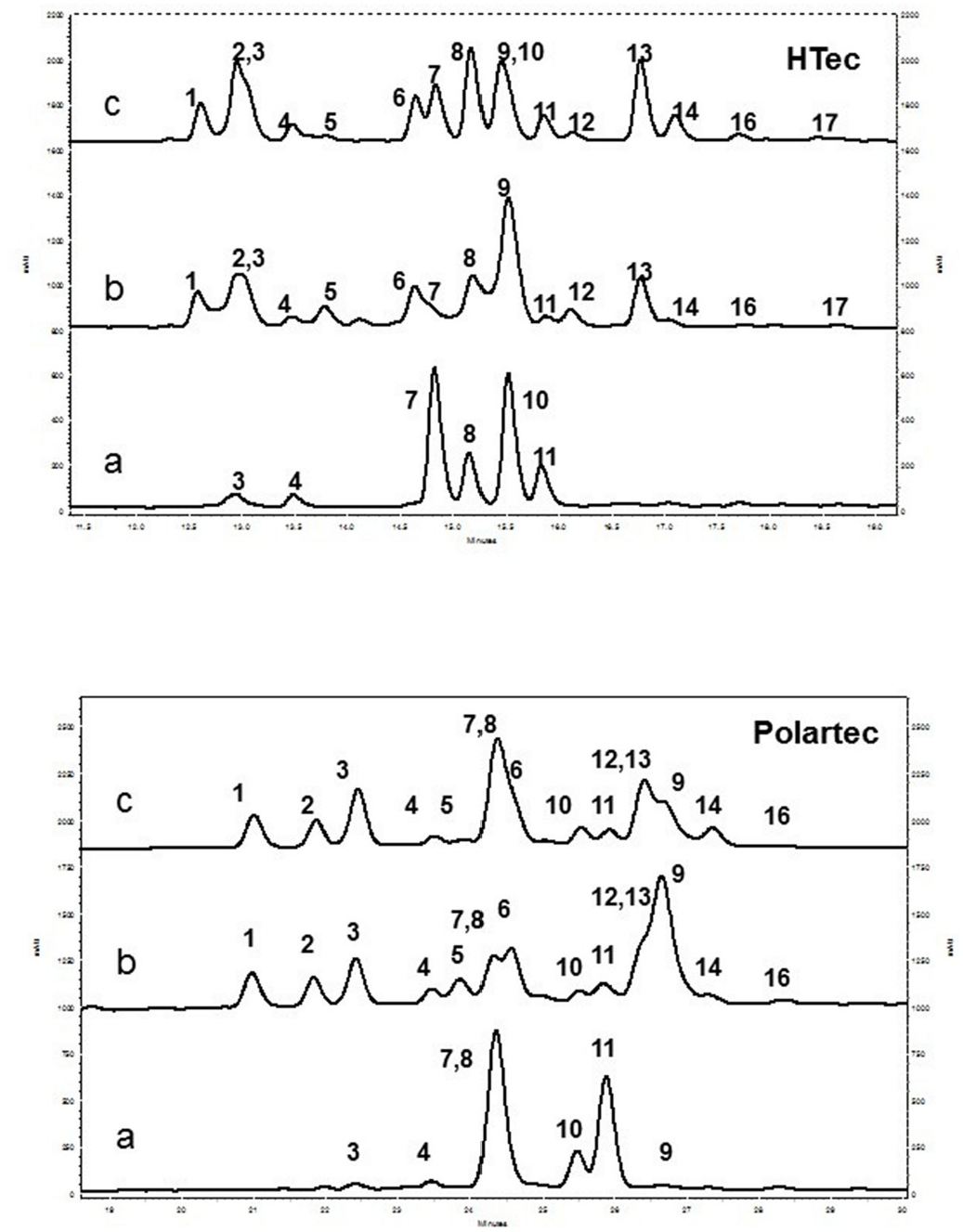
HPLC-DAD stand alone and coupled with LC/MS were used to determine the phenolic profile of the three species. The crude extracts were subjected to two different columns (Polartec and HTec), which provided a check for co-elution problems. While HTec is a classical C18 hydrophobic phase, the Polartec column is a C18 phase with embedded amido group leading to pronounced hydrophilic properties and separation mechanisms based both on hydrophobic (van der Waals) and polar interactions. The chromatograms obtained with the two columns are depicted in Fig. 2 View Fig . Examination of the HPLC profiles revealed important qualitative and quantitative differences in the phenolic composition of the two parental species S. maritima and S. alterniflora , while S. anglica indicated substantial similarities with S. alterniflora . A total of sixteen peaks were detected for S. alterniflora and S. anglica and only seven peaks for S. maritima ( Fig. 2 View Fig ). The hydrophilic properties of the Polartec column allowed the separation of peaks 1, 2, 3, which were in part co-eluted on HTec, and also of peak 9. The HTec column allowed a perfect separation of peaks 7 and 8, which were co-eluted on Polartec, and of peaks 10 and 11, which were poorly separated on Polartec. Compounds were identified from retention times, mass spectra, on-line UV spectra and comparison to standard when available.
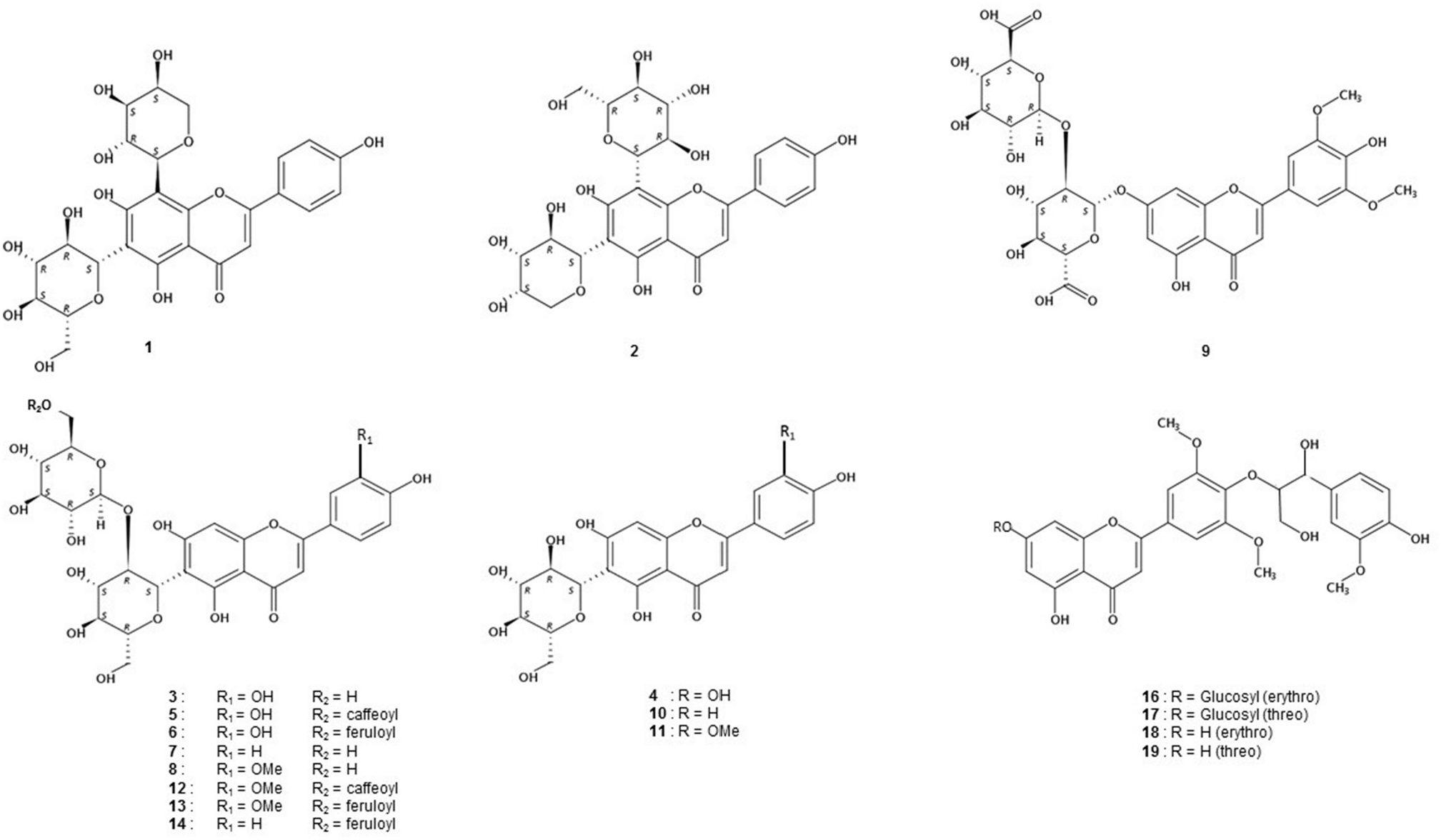
The three phenolic profiles were largely dominated by flavonoids. Comparison of retention time and UV spectra with those of reference standards indicated the absence of flavonols and flavones aglycones, except traces of tricin identified in the three species by comparison with an authentic standard (full list of standards used is given in Table S1 View Table 1 , and on-line UV spectra in Fig. S1 View Fig ; supplementary materials). Structural assignments were achieved by direct comparison with standards (compounds 4 and 10), or with previously positively identified compounds from S. maritima (compounds 7–8, 11; Grignon-Dubois and Rezzonico, 2019). Repeated analysis of the samples following acid hydrolysis permitted positive identification of compounds 1–8 and 10–14 as flavone C-glycosides and compounds 9 and 16 as flavone O - glycosides. Peaks 4, 7, 8, 10, 11 were respectively identified as isoorientin, isovitexin 2″- O -glucoside, isoscoparin 2″- O -glucoside, isovitexin and isoscoparin ( Fig. 3 View Fig , Tables S2–S View Table 2 3 View Table 3 ).
No known copyright restrictions apply. See Agosti, D., Egloff, W., 2009. Taxonomic information exchange and copyright: the Plazi approach. BMC Research Notes 2009, 2:53 for further explanation.
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |