Tsitsikamma Samaai and Kelly, 2002
|
publication ID |
https://doi.org/10.11646/zootaxa.4896.3.4 |
|
publication LSID |
lsid:zoobank.org:pub:FD238C7C-E3F8-408B-9711-9A0BFFF69692 |
|
DOI |
https://doi.org/10.5281/zenodo.4391108 |
|
persistent identifier |
https://treatment.plazi.org/id/0A5787DE-A978-FFC2-FF5C-6A80FDAD9210 |
|
treatment provided by |
Plazi |
|
scientific name |
Tsitsikamma Samaai and Kelly, 2002 |
| status |
|
Genus Tsitsikamma Samaai and Kelly, 2002 View in CoL
Type species. Tsitsikamma favus Samaai and Kelly, 2002 View in CoL , pg. 718, fig. 6A–G.
Diagnosis. Hemispherical, pedunculate, or encrusting Latrunculiidae with a smooth surface, sometimes folded, covered with large cylindrical or volcano-shaped oscules and raised fungiform areolate porefields. Colour in life is brown, dark liver brown, green, dark turquoise or pinkish. Texture is either extraordinarily tough and leathery or soft and compressible. Megascleres are anisostyles, thickest centrally, often slightly irregular and wavy; microscleres are isochiadiscorhabds, with typically three substructures being cylindrical-conical tubercles. A choanosome permeated with rigid honeycomb-like chambers visible to the unaided eye, surrounding a much softer interior with wispy tracts, or a single purse-like (sac-like) chamber with softer interior. Microscleres are present in an irregular palisade on the surface ectosome and lining the internal tracts. Ectosomal skeleton consists of tangential layer of anisostyles. The morphology of the cylindrical-conical tubercles is diverse. Chemistry includes pyrroloquinoline alkaloids (tsitsikammamines), and the discorhabdin derivatives 14-Bromo-3-dihydrodiscorhabdin C and 14-Bromodiscorhabdin C. The genus is endemic to the Agulhas ecoregion of South Africa and restricted to cold upwelling areas on the south and southeast coasts (modified from Samaai and Kelly 2002; Samaai et al. 2003).
Remarks. Parker-Nance et al. (2019) proposed two morphological groups, “ favus ” and “ pedunculata ”, for the genus Tsitsikamma , based on growth form and analysis of 28S rRNA sequences. We hereby propose the establishment of two new subgenera, Tsitsikamma ( Tsitsikamma) subgen. nov. Samaai & Kelly, 2002, and T. ( Clavicaulis ) subgen. nov., for the type species T. favus and T. pedunculata , respectively, based on the two very different morphologies displayed by these two species. Species in the nominotypical subgenus Tsitsikamma , containing the type species T. favus , have a thick encrusting to hemispherical growth form, a rigid honeycombed internal structure composed of dense spicule tracts supporting the choanosome, and large, thick microscleres with three (or four) whorls. Species in the new subgenus Clavicaulis , with T. pedunculata as type, have a purse or sac-like morphology, with a short thick stalk, a choanosome made up of a single chamber and with small, stout microscleres with two or three whorls.
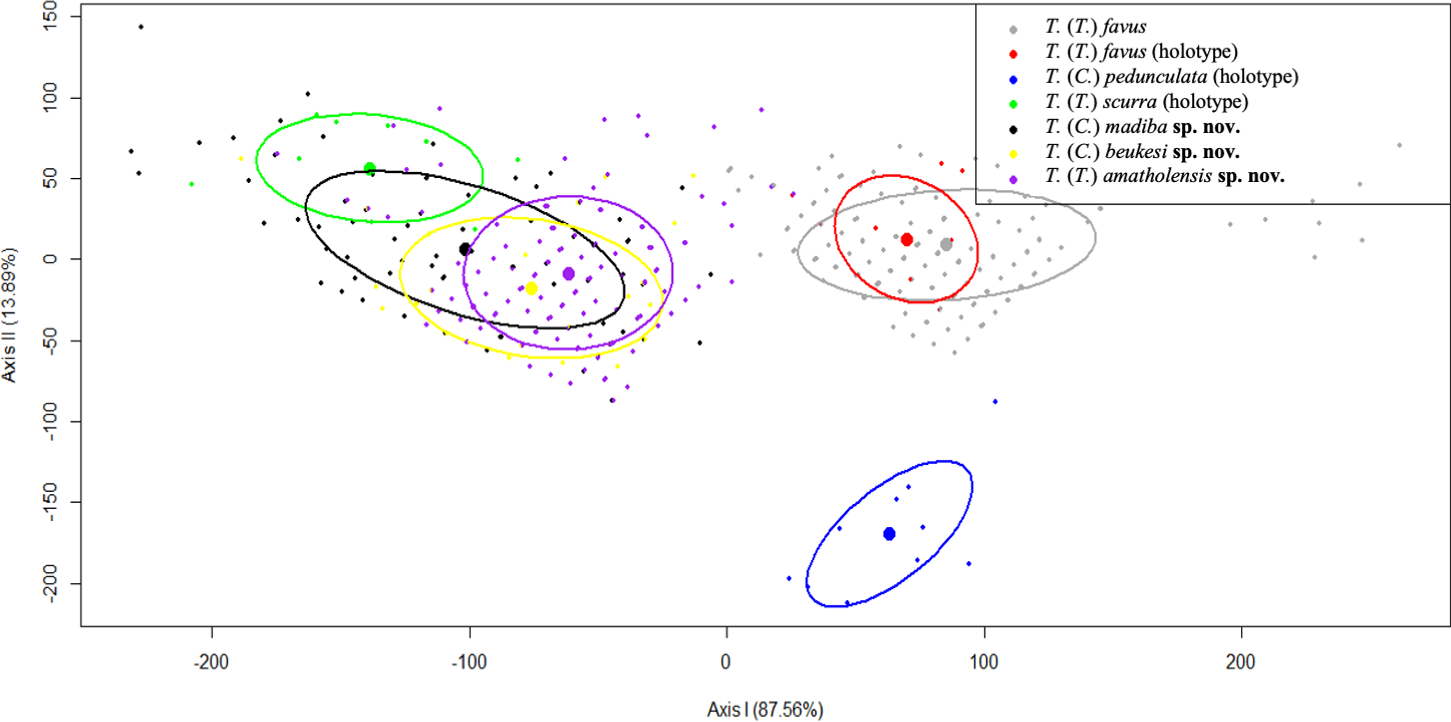
A multivariate analysis of spicule measurements (anisostyle length, discorhabd length, shaft and whorl length), was used to analyse the relative contribution of each morphometric variable in the distinction of the proposed new species and subgenera. Despite the obvious species-level differences in gross morphology, the use of morphometric variation, as an estimator of species distinction, was not able to separate the proposed new species, but separated T. favus , T. pedunculata and T. scurra Samaai, Gibbons, Kelly & Davies-Coleman, 2003 from each other ( Fig. 4 View FIGURE 4 ).
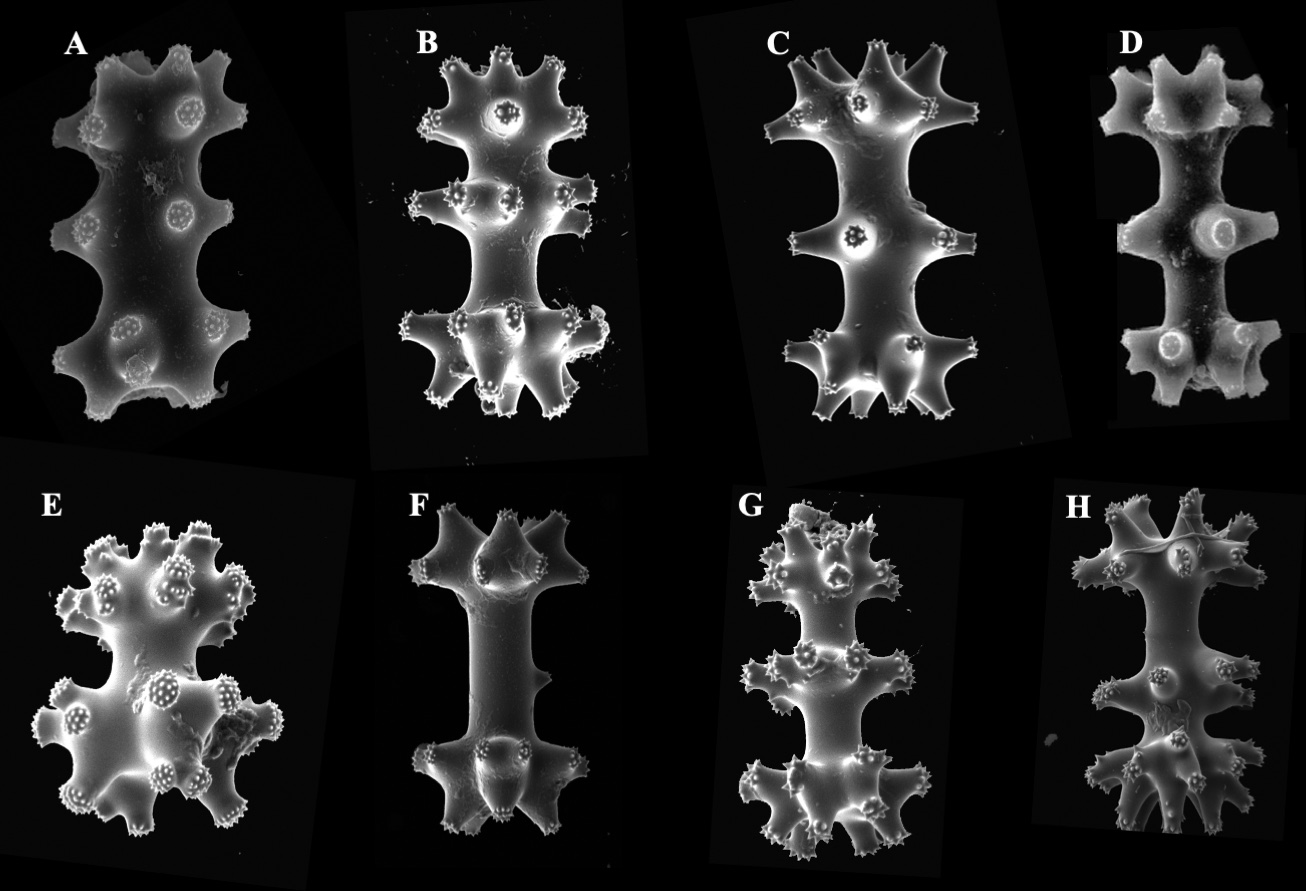
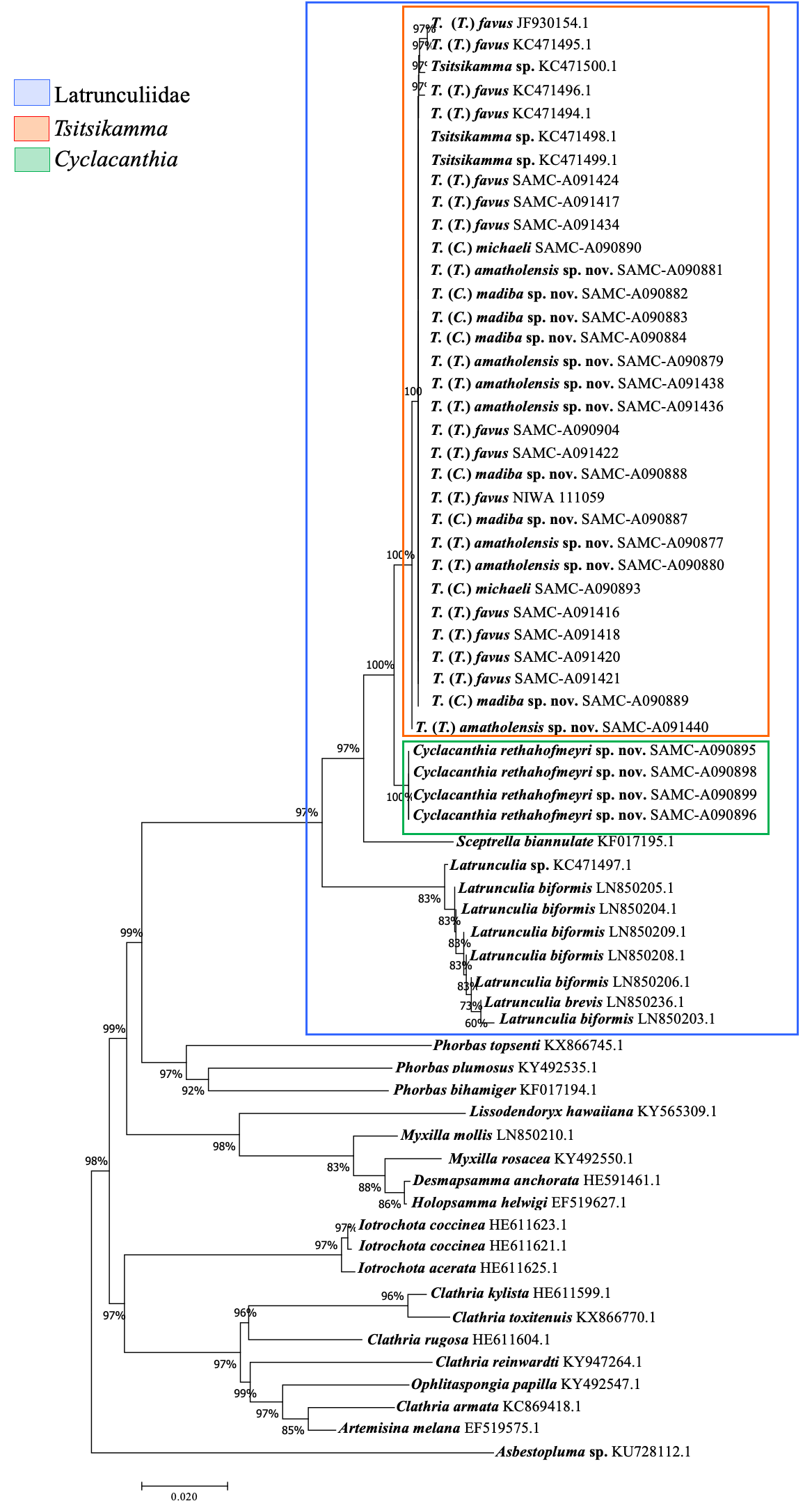
Similarly, DNA barcoding of the mitochondrial COI and the nuclear ITS of Tsitsikamma specimens, failed to clearly differentiate between specimens, despite clear morphological differences ( Figs 5 View FIGURE 5 , 6 View FIGURE 6 , 7 View FIGURE 7 ), a finding also obtained by Parker-Nance et al. (2019) based on COI and 28S rRNA gene sequences. The Tsitsikamma specimens all had the same COI haplotype, which clustered among previously published sequences of Tsitsikamma ( Fig. 6 View FIGURE 6 ). All specimens were grouped together into one cluster irrespective of whether they were stalked, oval, semispherical, rigid/soft, sac-/purse- or honeycomb-like or possessed two or three whorls of isochiadiscorhabd microscleres. No intra-specimen variation was found for the COI partition for the Tsitsikamma specimens, with the notable exception of one specimen, Tsitsikamma SAMC-A091440. This is rather surprising because COI sequences are considered variable and quite informative for population genetics and phylogeographic studies of demosponges (LópezLegentil et al. 2009; Reveillaud et al. 2011; Carella et al. 2016).
The high genetic similarity raises the question of how far these sponges are phylogenetically separated. The genetic results could also imply that the Tsitsikamma sponges included here represent one single species. Notwithstanding the above, strictly identical COI sequences have been reported for other sponges, e.g. Antarctic species of Rossella Carter, 1872 , Cinachyra Sollas, 1886 , Antarctotetilla Carella, Agell, Cárdenas & Uriz, 2016 and Siberian freshwater sponges. These studies showed that the mitochondrial COI M1-M6 partition failed to separate species within these cold-water genera, despite clear morphological differences (Schr̂der et al. 2003; Carella et al. 2016; Vargas et al. 2017). Vargas et al. (2015) also observed instances where DNA barcoding (COI) could not correctly assign a specimen to its species because multiple species share identical or near identical barcodes (e.g. Latrunculia , Artemisina Vosmaer, 1885 , Acanthorhabdus Burton, 1929 , Iophon Gray, 1867 ). In this study, as in the case for Latrunculia , Rossella and Iophon , Tsitsikamma is a cold-water genus and share identical barcode sequences, despite clear morphological differences (habit, isochiadiscorhabd morphology ( Fig. 5 View FIGURE 5 ), etc.).
The ITS sequences for Tsitsikamma ( Fig. 7 View FIGURE 7 ) were also uninformative and did not discriminate between the proposed morphological species (see Table 1; Fig. 5 View FIGURE 5 ). This was expected because of the slow evolving nature of this partition. The ITS tree recovered was different to the COI tree. The findings highlight the limitations of the morphometric measurements and gene sequences for distinguishing between specimens and the importance of rigorous morphological examinations of specimens within the family Latrunculiidae . The results may also indicate contrast-ing evolutionary rates between sponges in the family Latrunculiidae as found in other Demospongiae groups ( Heim et al. 2006). As for Antarctic Tetillidae Sollas, 1886 ( Carella et al. 2016), our results suggest either a particularly slow genetic evolutionary rate of the COI and ITS markers or a recent radiation with phenotypic characters evolving faster than the genetic markers studied. Until more data become available (e.g., further studies on cold-water Demospongiae families/genera vs. temperate/tropical families/genera) we prefer to maintain the proposed new species as separate due to the clear differences in morphology and isochiadiscorhabd structure, habit and distribution.
The COI partition was informative enough to confirm the monophyly of family Latrunculiidae and the relationships between the various genera in the family ( Fig. 6 View FIGURE 6 ). Our results concur with Parker-Nance et al. (2019) in supporting the monophyly of Latrunculiidae .
Based on morphological comparisons (habit, isochiadiscorhabd morphology, etc.) we describe three new species of Tsitsikamma within the two new subgenera: T. ( T.) amatholensis sp. nov.; T. ( C.) madiba subgen. et sp. nov.; T. ( C.) beukesi subgen. et sp. nov.
No known copyright restrictions apply. See Agosti, D., Egloff, W., 2009. Taxonomic information exchange and copyright: the Plazi approach. BMC Research Notes 2009, 2:53 for further explanation.
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
SubClass |
Heteroscleromorpha |
|
Order |
|
|
Family |