Ambrosia artemisiifolia
|
publication ID |
https://doi.org/10.1016/j.phytochem.2014.10.022 |
|
DOI |
https://doi.org/10.5281/zenodo.10522626 |
|
persistent identifier |
https://treatment.plazi.org/id/304587B9-0E68-4206-9A35-FD52FF18F907 |
|
treatment provided by |
Felipe |
|
scientific name |
Ambrosia artemisiifolia |
| status |
|
2.1. Phenolics and flavonoid concentration in the pollen and SPP extracts of A. artemisiifolia L View in CoL
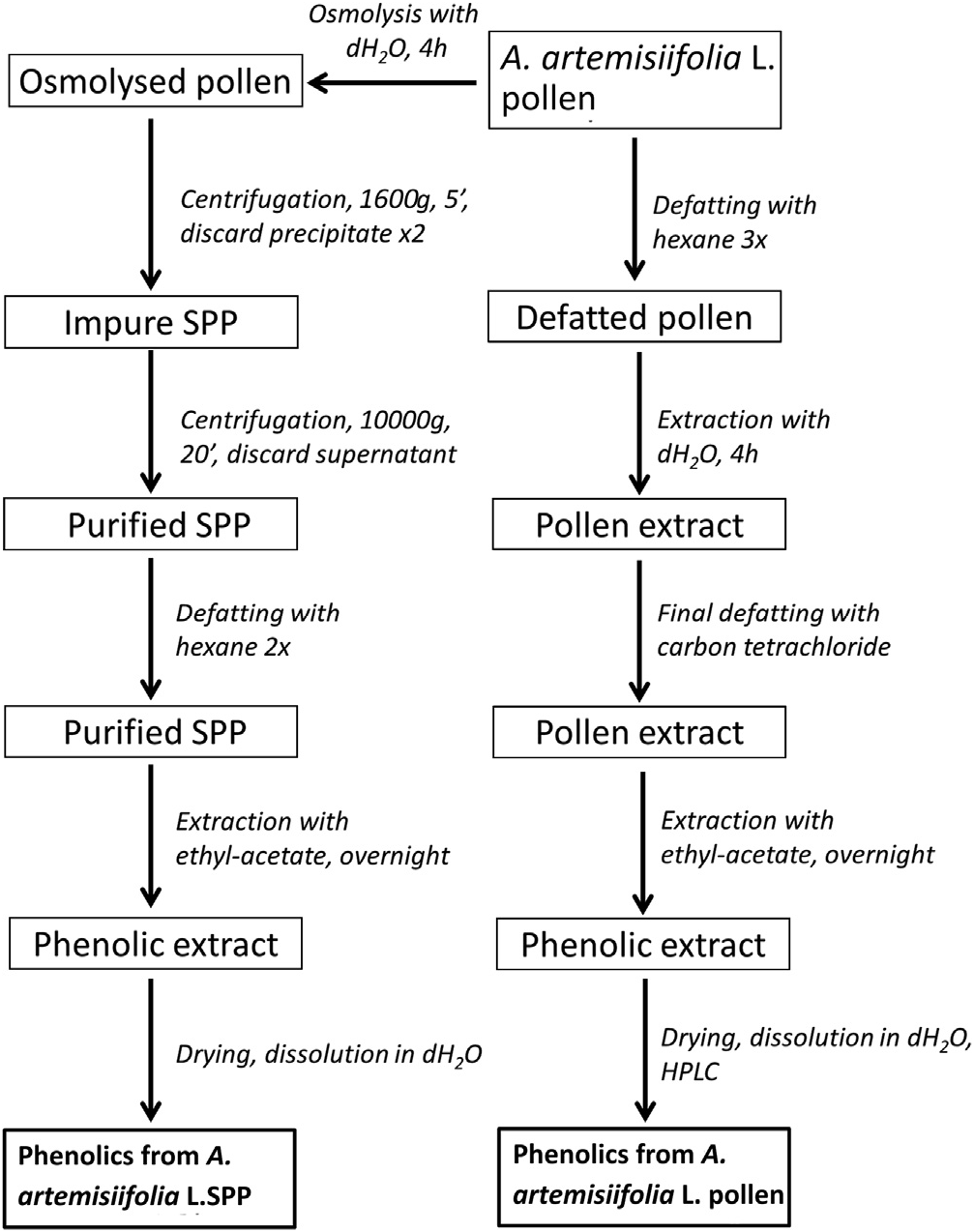
Separation scheme for extraction of phenolic compounds is given in Fig. 1 View Fig . It was determined that the ethanol-based extract of A. artemisiifolia L. pollen contains phenolics at a concentration of 6.79 ± 0.05 mg GAE/g pollen, while its SPP value is 4.56 ± 0.06 mg GAE/g SPP, being 33% significantly lower from the whole pollen grain extract. In addition, flavonoid content in these extracts mounted up to 98% (4.45 ± 0.01 mg /g) of total phenolics in SPP and 70% for whole pollen extract (4.71 ± 0.04 mg /g). This is in accordance with structural data ( Tables 1 View Table 1 and 2 View Table 2 ).
Water-soluble phenolics are present at 4.51 ± 0.02 mg GAE/g pollen. This value of phenolics in A. artemisiifolia L. pollen is significantly lower than the value previously reported for the phenolic content of A. artemisiifolia L. herb, which can go as high as 4.35% of total dry weight ( 43.5 mg /g dry herb) ( Maksimovic, 2008). This can be due to the different roles phenolics might play in pollen vs. plant leaves and stalks ( Falasca et al., 2010). The amount of flavonoids obtained from the aqueous pollen extract is 3.95 ± 0.24 mg /g pollen, which is 87.5% of total soluble phenolic content. It can be concluded that the flavonoids are the main components of pollen and SPP phenolic extracts, as opposed to dry plant flowering summits, where they make up less than 5% of total phenolics content ( Maksimovic, 2008). In addition, values obtained for water- and ethanol extracts in A. artemisiifolia L. pollen are also lower than majority of bee pollen (flowering) species, while results on sub-pollen particle phenolics amount are novel and thus incomparable to literature data. However it is noticeable that SPP have one-third lower content of the total phenolics than the whole pollen grains, which could be important when concerned with allergy modulating properties of polyphenols. It is known that beside direct anti-allergic effect ( Yoon et al., 2006), polyphenolic compounds could alleviate allergy by cross-linking with allergens and thus reducing their allergenicity ( Tantoush et al., 2011).
2.2. Antioxidative properties of the pollen and SPP extract
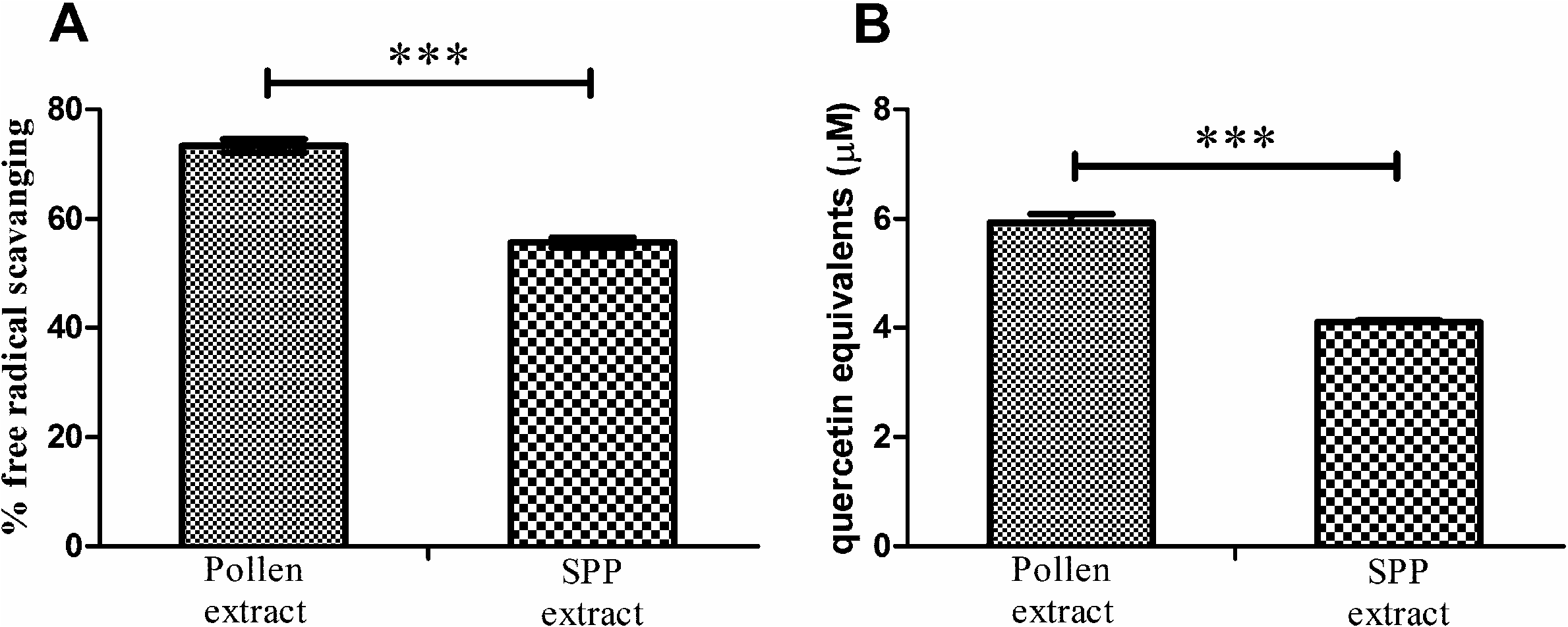
DPPH assay is commonly used to assess free radical scavenging of a compound or a mixture. Ambrosia pollen extract showed 73 ± 2% free radical scavenging, as compared to a methanol blank. This value is equivalent to the scavenging activity of 28.4 µM butylated hydroxyl toluene (BHT) standard. SPP extract exhibited lower activity, with 56 ± 2% of free radical scavenging activity (23% decrease). The differences are statistically significant ( p <0.001) ( Fig. 2A View Fig ). Both values can be considered as significant free radical scavenging activity and are corroborated by papers by other authors ( Negri et al., 2011).
ABTS+ assay was used to measure the antioxidative ability of the extracts. Ambrosia pollen extract showed activity equivalent to 5.9 + 0.2 µM quercetin, while the activity of SPP extract was significantly lower ( p <0.001) at 4.1 + 0.1 µM quercetin equivalent (31% decrease). The results are shown in Fig. 2B View Fig . Both values are lower than expected from the previously obtained values of pollen phenolics concentration. Although quercetin was used as a standard due to its structural similarity to main components of the mixture, most of the phenolics present in the extract are glycosylated or otherwise modified, which can affect the antioxidative capacity of the mixture.
2.3. Characterization of A. artemisiifolia L. pollen water-soluble phenolics on Orbitrap LTQ XL
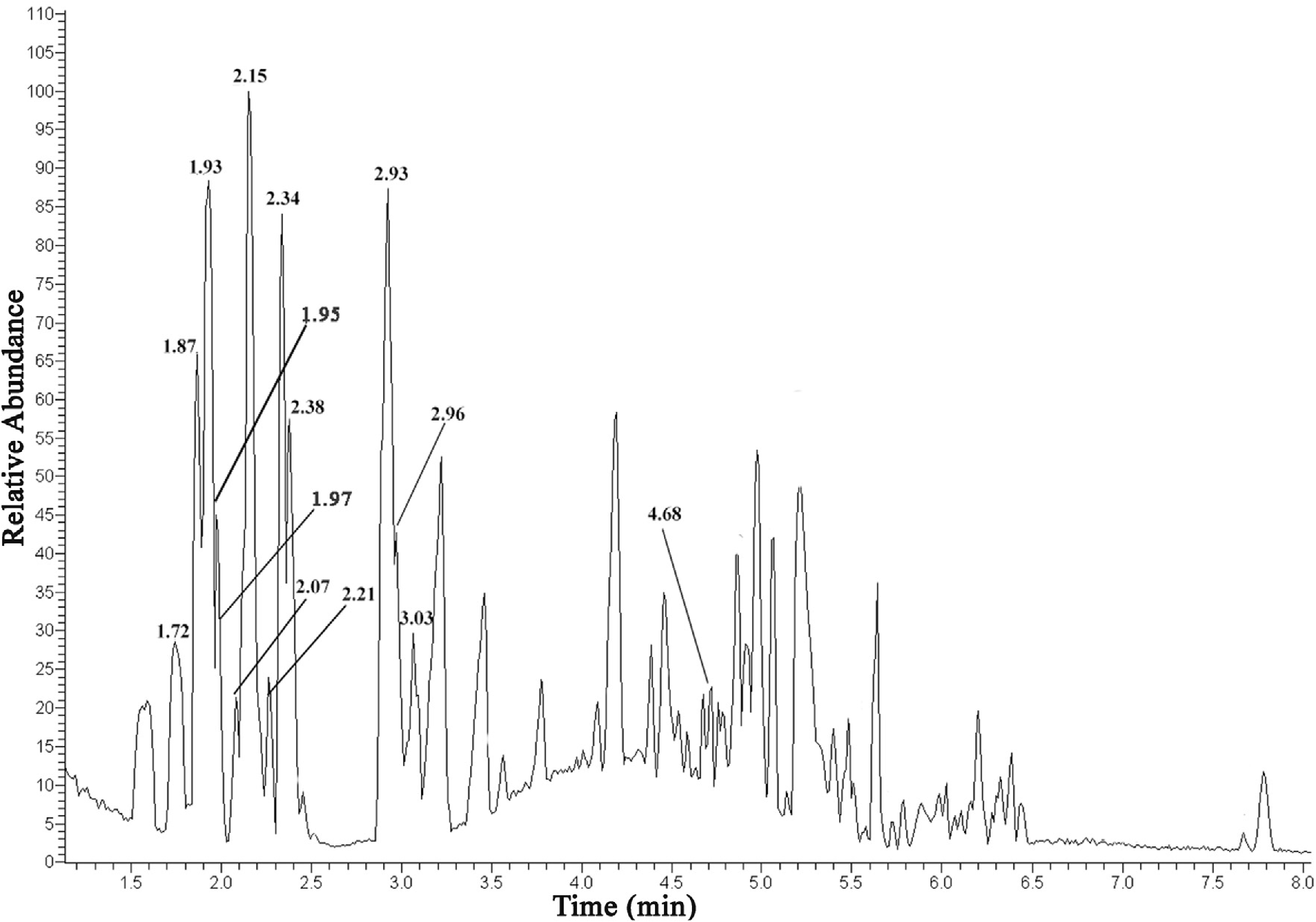
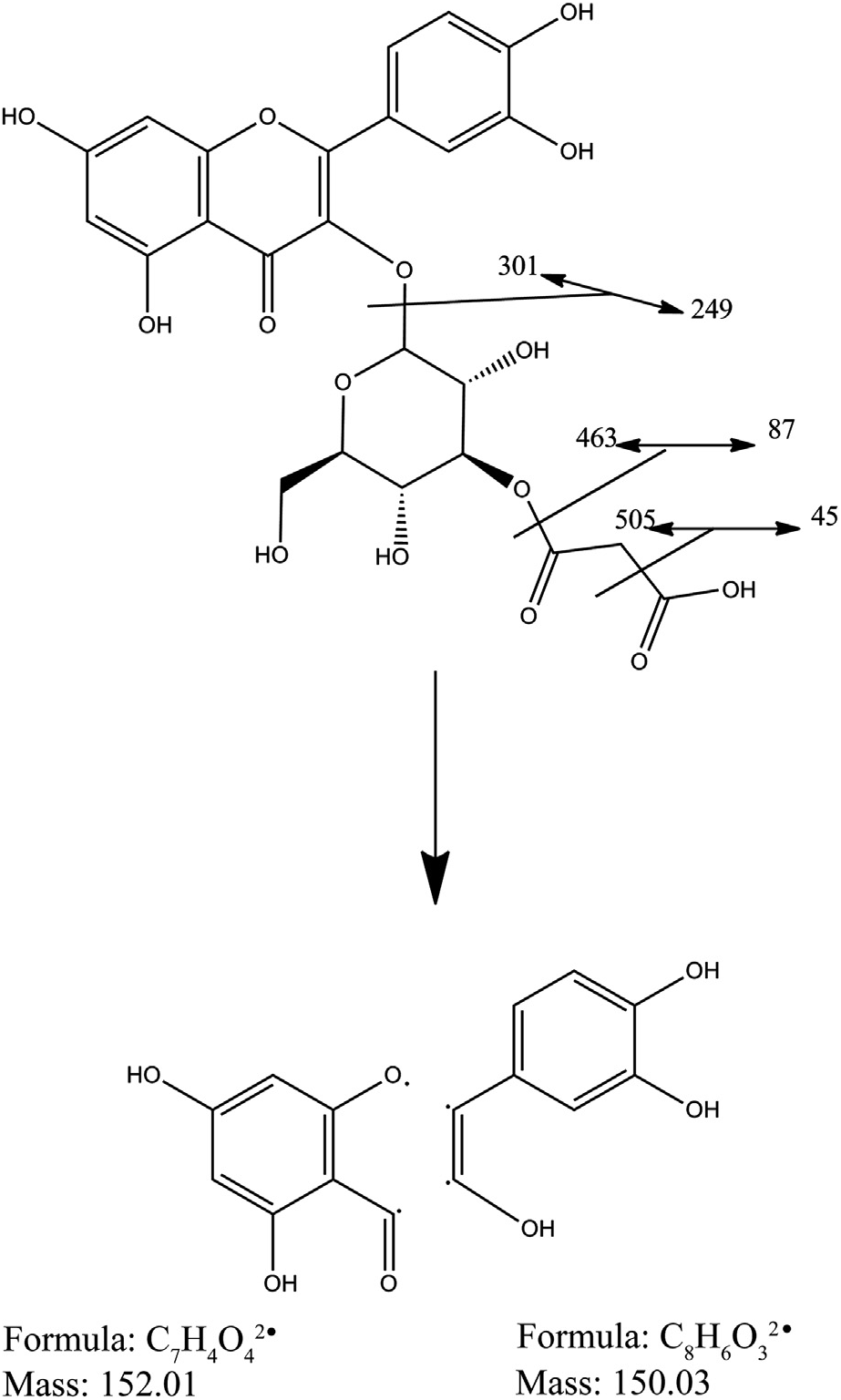
An Orbitrap analysis showed that A. artemisiifolia L. pollen phenolics have a complex and diverse composition, including phenolic derivates of polyamide spermidine, which we were unable to detect with the triple quadrupole detector (data not shown). All of the phenolic glycosides detected with high resolution and sensitive UHPLC /ESI-LTQ-MS– MS Orbitrap, including m/z higher than 570 (di- and tri-glycosides of isorhamnetin), are shown Table 1 View Table 1 and its base peak chromatogram is shown in Fig. 3 View Fig .
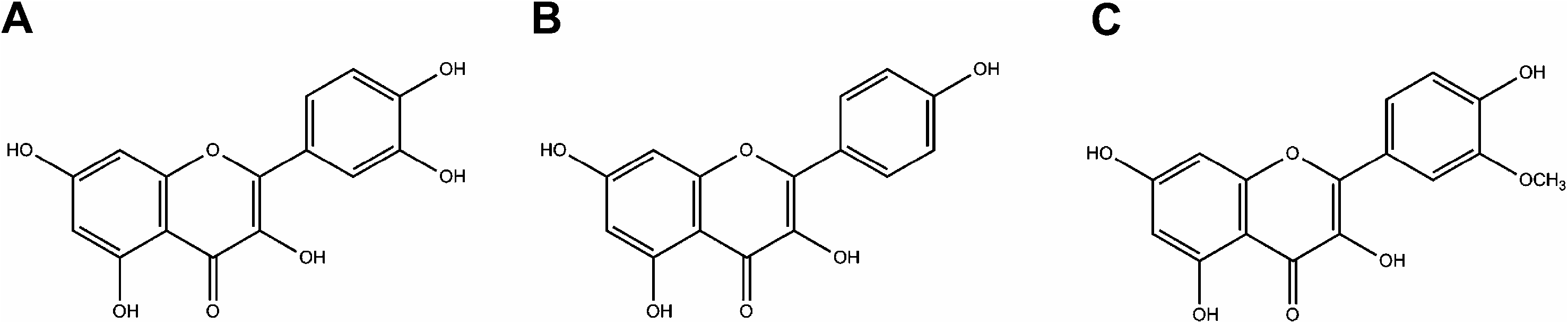
All detected glycosides are derivatives of three main flavonols: quercetin, O -methylquercetin (isorhamnetin), and kaempferol ( Fig. 4 View Fig ). For each aglycone, a series of peaks (satellite sets) was observed, representing unsubstituted and malonylated hexosides and hexuronides.
Fragment analysis allowed identification of [Y 0 — 2 H — CO] — ions, a characteristic ion-peak of 3-O-monoglucosides, confirming the structures proposed with preliminary results ( Vukics and Guttman, 2010).
In addition to the aglycone (Y 0 —) fragment, an abundant radical aglycone [Y 0 — H] —. product ion is also formed, indicating a loss of 3- O-glucosyl fragment (all fragments with m / z of aglycone 285, 301, 315 accompanied by ions at masses 284, 300, 314), further proving that mostly 3-O-glycosilated fragments are present in the mixture ( Hvattum and Ekeberg, 2003). Fragmentation of the flavonol derivative quercetin-3-glucoside-3-malonate is shown in Fig. 5 View Fig .
The abundance ratio of the radical aglycone to the regular aglycone product ion originating from cleavage at the 3- O glycosidic bond increases with increasing OH substitution on the B-ring, whereas the opposite holds for 7-O-glycosides ( March et al., 2006). This phenomenon can be observed when comparing kaempferol and quercetin derivatives (peaks at retention time (RT) 1.72 and 2.09, as well as 1.89 and 2.09). Aglycone identities were confirmed by following 1,3 B 0 and 1,3 A 0 fragments present in the MS 2 spectra. Both quercetin and isorhamnetin can produce 1,3 A 0 fragment with m / z of 151 in negative mode, in a reaction that proceeds via retro Diels–Alder mechanism (RDA). These compounds were distinguished according to their molecular ions and Y 0 — 2 H — CO ions.
Compounds having a methoxy substituent show relatively weaker RDA fragmentation, observable when comparing the fragmentation spectra of quercetin and isorhamnetin derivatives (RT 2.2 and 2.21, 1.72 and 1.93) ( Fabre et al., 2001).
2.4. Spermidine derivatives in the pollen extract of A. artemisiifolia View in CoL L
Phenolamides constitute a diverse and quantitatively major group of secondary metabolites resulting from the conjugation of a phenolic moiety with polyamides, such as spermidine ( Bassard et al., 2010). Several spermidine amide derivatives have been detected in the A. artemisiifolia L. pollen extract. Among them, the most prominent were di-coumaroyl-caffeoyl and tri-coumaroyl containing compounds. Fragmentation patterns for spermidine derivatives are shown in Fig. 6 View Fig .
Peak at retention time 2.93, with m / z of the [M — H] — ion of 598.2573 showed characteristic fragmentation pattern ( Sobolev et al., 2008) with fragments at m / z 478, attributed to the loss of 120 Da fragment from p -coumaric residue ([M — H] — A HO A C6 A H4 A CH @ CH), m / z 436 (loss of coumaroyl residue) and m / z 358 (loss of 120 Da fragment at N 5 and breakage of C A C bond of the caffeoyl residue linked at N 10). The compound was identified as di-coumaroyl caffeoyl spermidine.
Observed masses can be explained by formation of homolytic cleavage/deprotonation products during fragmentation. Few papers have been published so far regarding the fragmentation of polyamide phenolics ( Narvaez-Cuenca et al., 2013). To the best of our knowledge, formation of radical products of spermidine derivatives in MS 2 has not been described so far.
Similarly, at 2.96 min retention time, a peak with m / z of 582.262 for the [M — H] — ion, with corresponding fragments in MS 2 at m / z 462, 436 and 358 has identical fragmentation mechanism as for the previous compound, with the only 16 Da difference for the m / z 462 and m / z 436 fragments. The difference stems from the replacement of a caffeoyl residue with a coumaroyl residue. The compound was identified as tri- p -coumaroyl spermidine.
Spermidine and other polyamide compounds are widely present in all organisms, known to affect many processes, in both plants ( Falasca et al., 2010), and animals ( Igarashi and Kashiwagi, 2010). They have been shown to function naturally as free radical scavengers ( Ha et al., 1998). Spermidine acyl derivatives such as its amides have already been reported in pollen ( Bassard et al., 2010; Kite et al., 2013). Recently, polyamides were reviewed by Bassard et al. (2010), who concluded that di- and tri-substituted hydroxy-cinnamoyl conjugates, particularly of spermidine and putrescine, are major metabolites of pollen and suggested to have an ecological role as defense compounds against viruses, bacteria, and fungi, and could deter herbivores from eating plants. The spermidine conjugates are implicated in protection against pathogens, detoxifying phenolic compounds, and/or serving as a reserve of polyamines that are available to actively proliferating tissues, although not always essential for survival.
In addition, polyamides and their derivatives play a regulatory role in several immunologic processes, including allergic reactions ( Bueb et al., 1991; Hoet and Nemery, 2000), regulation of T cell function, cell migration and growth in local inflammation ( Ferioli et al., 2000).
| L |
Nationaal Herbarium Nederland, Leiden University branch |
| MS |
Herbarium Messanaensis, Università di Messina |
| O |
Botanical Museum - University of Oslo |
| H |
University of Helsinki |
| B |
Botanischer Garten und Botanisches Museum Berlin-Dahlem, Zentraleinrichtung der Freien Universitaet |
| A |
Harvard University - Arnold Arboretum |
No known copyright restrictions apply. See Agosti, D., Egloff, W., 2009. Taxonomic information exchange and copyright: the Plazi approach. BMC Research Notes 2009, 2:53 for further explanation.
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |