Palaeoagraecia brunnea Ingrisch, 1998
|
publication ID |
https://doi.org/10.11646/zootaxa.5072.3.2 |
|
publication LSID |
lsid:zoobank.org:pub:441034FD-431E-43AA-B318-49078C4DABB5 |
|
DOI |
https://doi.org/10.5281/zenodo.5747937 |
|
persistent identifier |
https://treatment.plazi.org/id/396087F6-FFF2-9F18-FF3D-023FFA90FCA6 |
|
treatment provided by |
Plazi |
|
scientific name |
Palaeoagraecia brunnea Ingrisch, 1998 |
| status |
|
18. Palaeoagraecia brunnea Ingrisch, 1998 View in CoL 布氏+ĸø
FIGURES 11–12 View FIGURE 11 View FIGURE 12
Material examined. 1 female, Mengjiao, Cangyuan , Yunnan, June 4, 2021, coll. by Jing Liu.
Distribution. Guangdong, Guangxi (Shiwandashan), Hainan, Yunnan (Cangyuan).
The complete mitogenome of Palaeoagraecia brunnea Ingrisch, 1998 was 15984 bp in length, including 13 protein-coding genes, 22 transfer RNA genes, 2 ribosomal RNA unit genes and a control region (A+T-rich regions). The arrangement of 37 genes was the same as other Conocephalinae species, among them, 9 protein-coding genes and 14 transfer RNA genes were located on the majority strand (J-strand), and the remaining genes (4 protein-coding genes, 8 transfer RNA genes and 2 ribosomal RNA unit genes) encoded on the minority strand (N-strand) ( Table 2 View TABLE 2 ). The nucleotide composition of the mitogenome (A=36.2%, T =34.0%, C=18.7%, G =11.1%) exhibited an obvious AT nucleotide bias, and the A + T content reached 70.2% ( Table 3 View TABLE 3 ). There were 14 gene overlapping regions and 19 intergenic spacer regions with a total length of 47 bp and 394 bp, respectively, which were less compact than other Conocephalinae mitogenomes. Notably, the longest intergenic spacer region in the mitogenome was found between ND 3 and trnA, with a length of 214 bp ( Table 2 View TABLE 2 ).
The total length of 13 protein-coding genes was 11150 bp. Most protein-coding genes started with the codon ATN ( ATA, ATG or ATT), except for COI gene that initiated with the codon CCG. Eleven protein-coding genes ended with typical stop codons TAA, and COII and ND 4 genes end with T ( AA) as the incomplete stop codons. The total length of 22 transfer RNA genes was 1464bp, and in size from 63 to 71 bp. The two ribosomal RNA unit genes were located between trnL1 and A+T-rich regions, separated by trnV, with the length of 1312 bp and 847 bp, respectively. The length of control region was 864 bp. A comprehensive analysis of nucleotide composition showed that the A + T content of control region was the highest in the whole mitogenome sequence at 78.8%. In addition, the A+ T content of 13 protein-coding genes was 68.5% and the A+ T content in the third codons (70.7%) was significantly higher than that in the first (66.8%) and second (67.9%) codons ( Table 3 View TABLE 3 ).
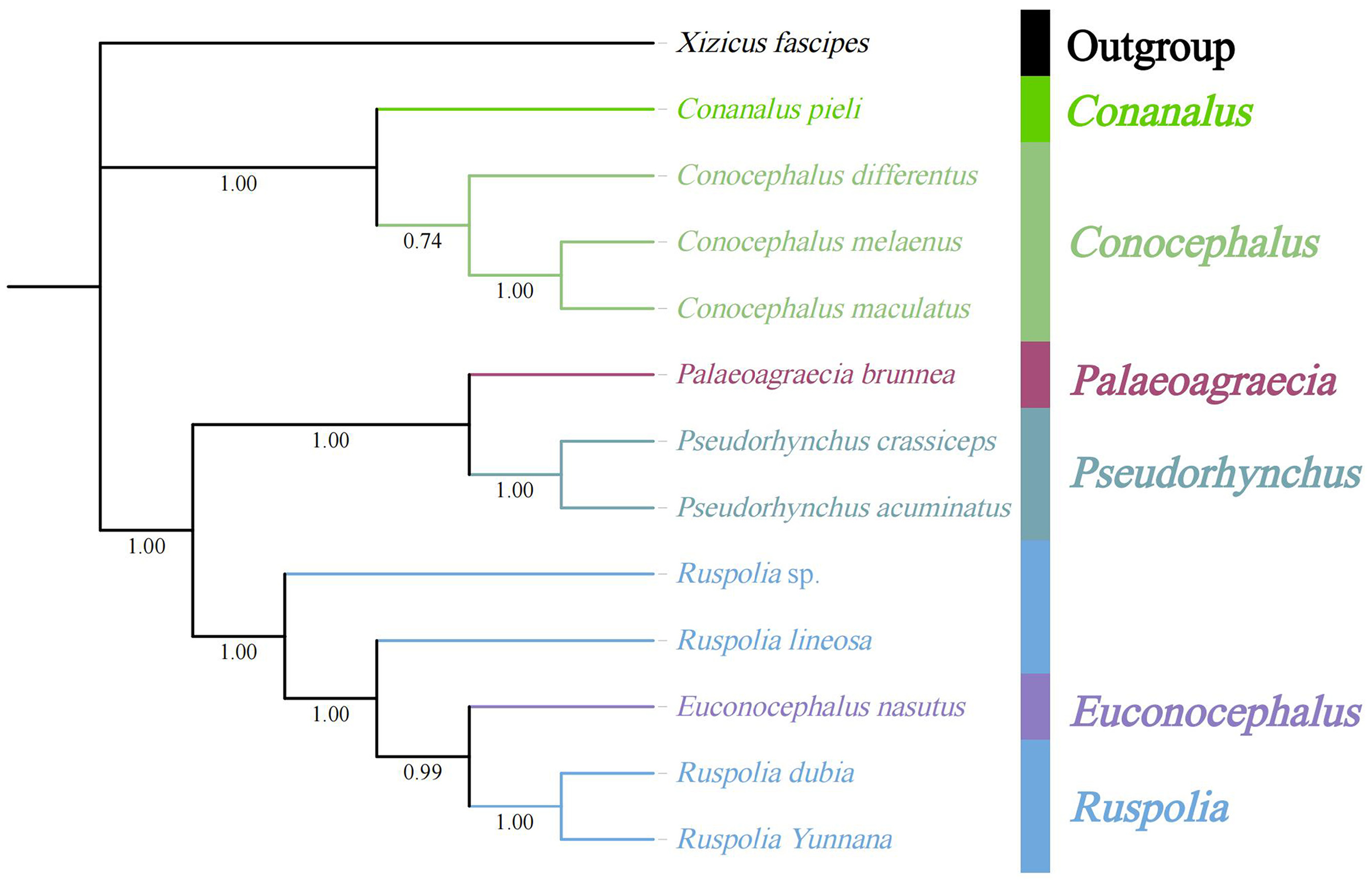
The phylogenetic tree showed that seven genera of Conocephalinae were divided into two clades with high nodal support values ( Fig. 13 View FIGURE 13 ). Apart from the outgroup, the tribe Conocephalini including the genus Conanalus and three species of Conocephalus were gathered together and found to be monophyletic. In the other main clade, the tribe Copiphorini was paraphyletic with Agraeciini nested within and the relationships between the genera were as follows: ( Pseudorhynchus +( Palaeoagraecia +( Anelytra +( Ruspolia + Euconocephalus )))) ( Fig. 13 View FIGURE 13 ). The phylogenetic relationships of the tribe Copiphorini recovered by the mitogenomes were not identical to the traditional morphological classification but consistent with the results of Muglestona et al. (2013) and Li et al. (2019).
| T |
Tavera, Department of Geology and Geophysics |
| ND |
University of Notre Dame |
| ATA |
Ataturk Universitesi |
| CCG |
Chengdu College of Geology |
| TAA |
Estonian University of Life Sciences |
| AA |
Ministry of Science, Academy of Sciences |
No known copyright restrictions apply. See Agosti, D., Egloff, W., 2009. Taxonomic information exchange and copyright: the Plazi approach. BMC Research Notes 2009, 2:53 for further explanation.
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
SubFamily |
Conocephalinae |
|
Tribe |
Agraeciini |
|
SubTribe |
Salomonina |
|
Genus |