Chromadoridae, Filipjev, 1917
|
publication ID |
https://doi.org/10.11646/zootaxa.4578.1.1 |
|
publication LSID |
lsid:zoobank.org:pub:CE27CD48-70BB-4979-9C67-A1A2E1BAAFFD |
|
DOI |
https://doi.org/10.5281/zenodo.5587324 |
|
persistent identifier |
https://treatment.plazi.org/id/52215C67-FFC5-FFB7-FF21-F92E8219F864 |
|
treatment provided by |
Plazi |
|
scientific name |
Chromadoridae |
| status |
|
Phylogeny of Chromadoridae
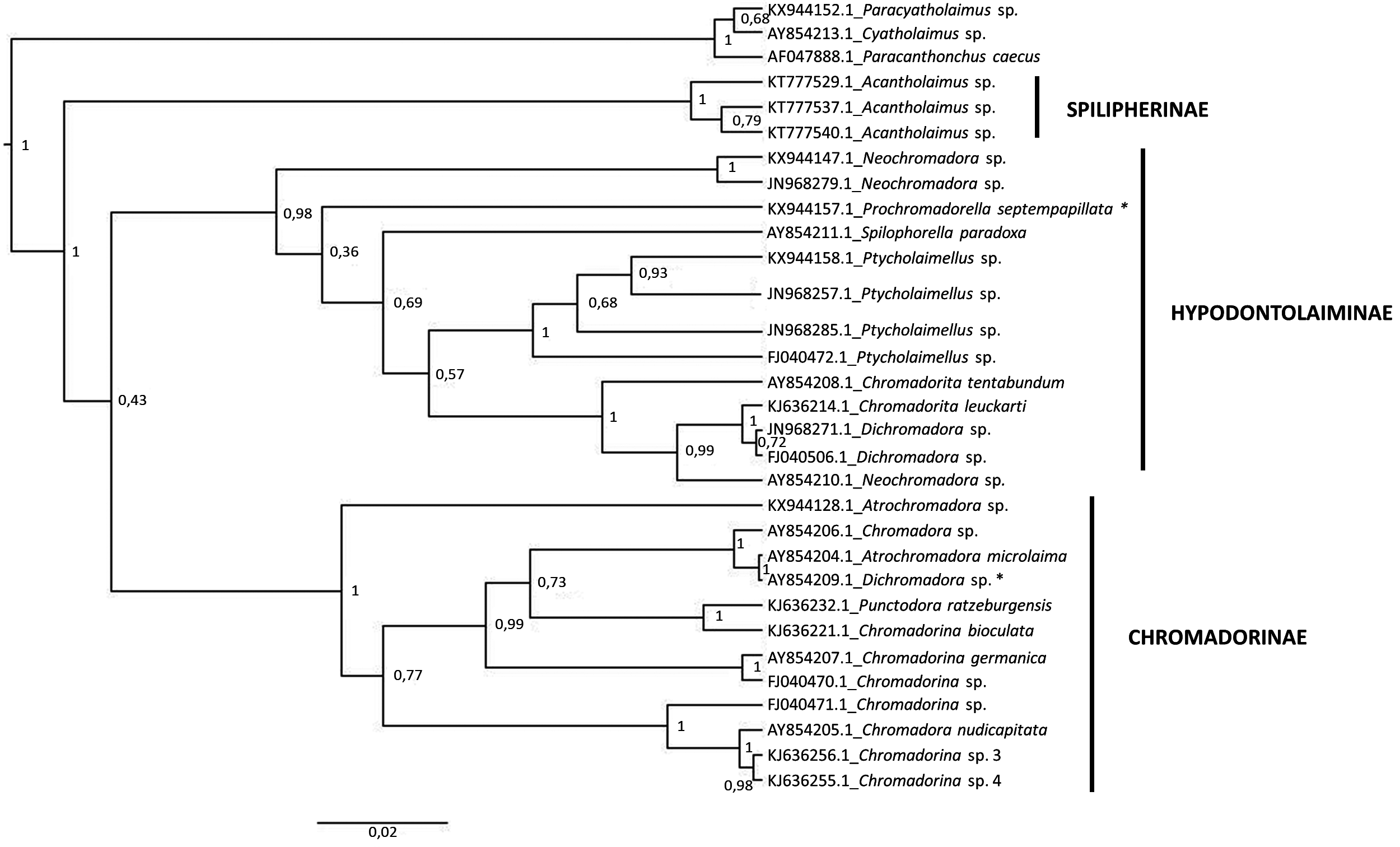
A total of 28 sequences of 18S rDNA were retrieved from the GenBank (Appendix 1). These sequences covered 11 genera, of which only nine were identified to species level. The 18S-based molecular phylogenetic tree recovered three monophyletic clades with high support ( Figure 6 View FIGURE 6 ), which correspond to the subfamilies Spilipherinae , Chromadorinae and Hypodontolaiminae . The sequence of Prochromadorella septempapillata Platt, 1973 , currently classified within the Chromadorinae was grouped with species of Hypodontolaiminae , and the inverse occurred with a Dichromadora sequence ( AY854209 View Materials ). The tree did not recover the monophyly of most genera. In the case of Chromadorita , C. tentadunbum , a misspelling of Chromadorita tentabunda de Man, 1890 , is considered as a complex Chromadorita / Innocuonema by Platt & Warwick (1988). The molecular data corroborated the hypothesis that this species does not belong to Chromadorita and may be classified as Innocuonema tentabunda (de Man 1890) . Chromadorina also showed a difficult taxonomy with many species synonymized or transferred from other genera (see discussion in the Taxonomic session). The high divergence between these sequences and their phylogenetic position suggests that this genus needs a careful re-evaluation. Sequences of Dichromadora , Neochromadora , Atrochromadora and Chromadora that were not clustered together with congeneric species probably are misidentified sequences in the GenBank. Problems of misidentification in genetic data banks are common across taxa ( Vilgalys 2003) and have a negative effect on the resolution of molecular phylogenies ( Holovachov 2016). The Dichromadora sequence ( AY854209 View Materials ), for example, is identical to the Atrochromadora microlaima sequence ( AY854204 View Materials ).
From 28S rDNA region, a total of 11 sequences were obtained covering eight genera and four nominal species (Appendix 1). Once again, the analyses recovered three monophyletic clades roughly corresponding to the three subfamilies ( Figure 7 View FIGURE 7 ). The sequences of Dichromadora and Chromadorina were grouped into Chromadorinae and Hypodontolaiminae , respectively, contradicting their current classification. This could be another case of misidentified sequences, since these sequences were not identified to species level.
Despite the absence of defined synapomorphies for the subfamilies, the molecular phylogenies were able to recover the classification recognized by Lorenzen (1994) and Tchesunov (2014). It is worth noting that the clade Spilipherinae was represented by a single genus and there are no sequences available for Euchromadorinae and Harpagonchinae . The addition of new data will clarify the systematics within the family. Along with this, the material deposited in the GenBank exemplifies how problematic it is the identification of Chromadoridae species. This is a common issue among marine nematodes and is a consequence of the poorly described species, large number of unknown species and scarcity of specialists. Validation of species lists (e.g. Venekey et al. 2014), integrative taxonomical descriptions (e.g. Cunha et al. 2013, Leduc et al. 2017), and systematics reviews (e.g. Miljutin & Miljutina 2016) are urgently needed within this group.
No known copyright restrictions apply. See Agosti, D., Egloff, W., 2009. Taxonomic information exchange and copyright: the Plazi approach. BMC Research Notes 2009, 2:53 for further explanation.
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
SubFamily |
Spilipherinae |
|
Genus |