Echiniscus
|
publication ID |
https://doi.org/ 10.1111/zoj.12090 |
|
persistent identifier |
https://treatment.plazi.org/id/666D485F-FFAE-FFE8-FEFC-F8B0FD095BC5 |
|
treatment provided by |
Marcus |
|
scientific name |
Echiniscus |
| status |
|
DESIGN IN ECHINISCUS View in CoL
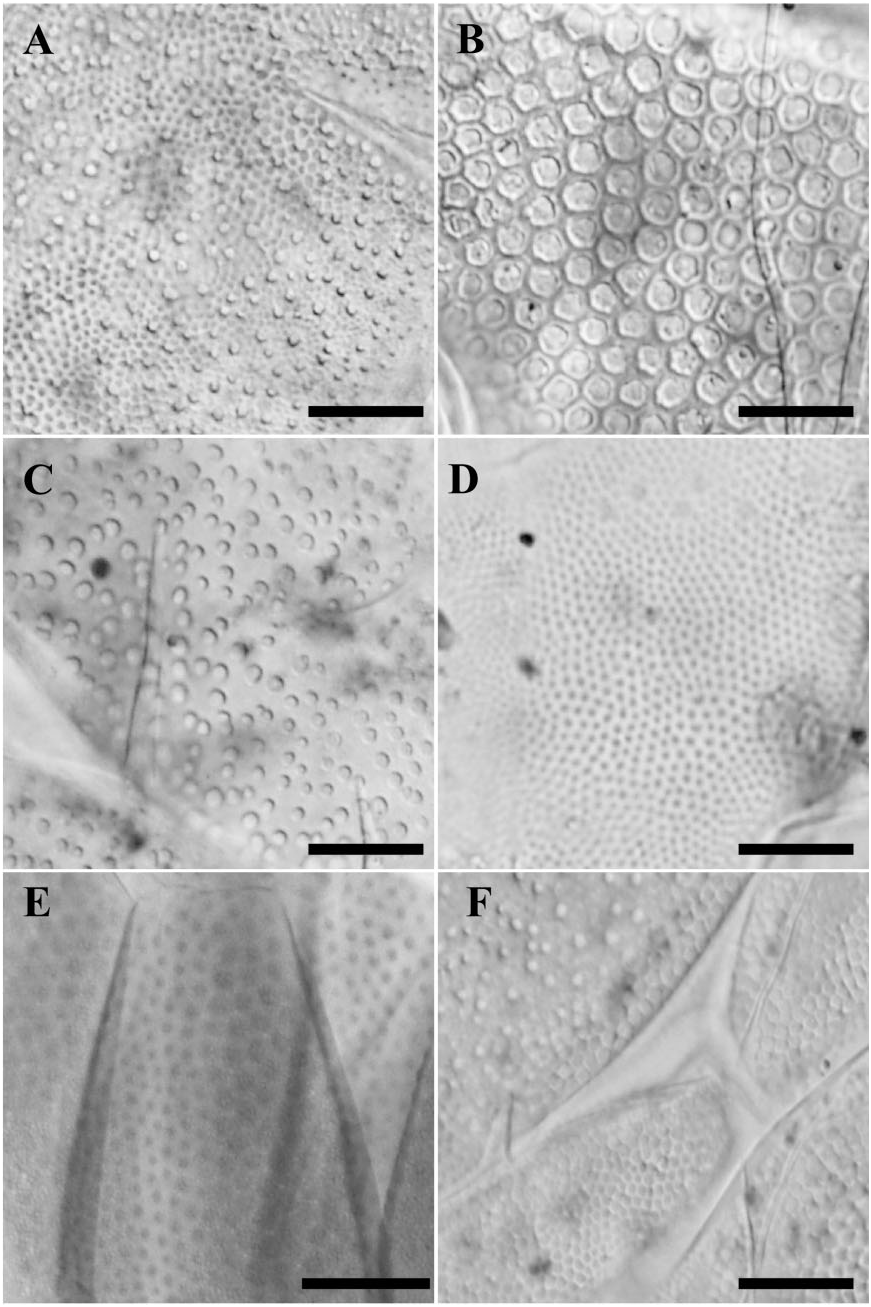
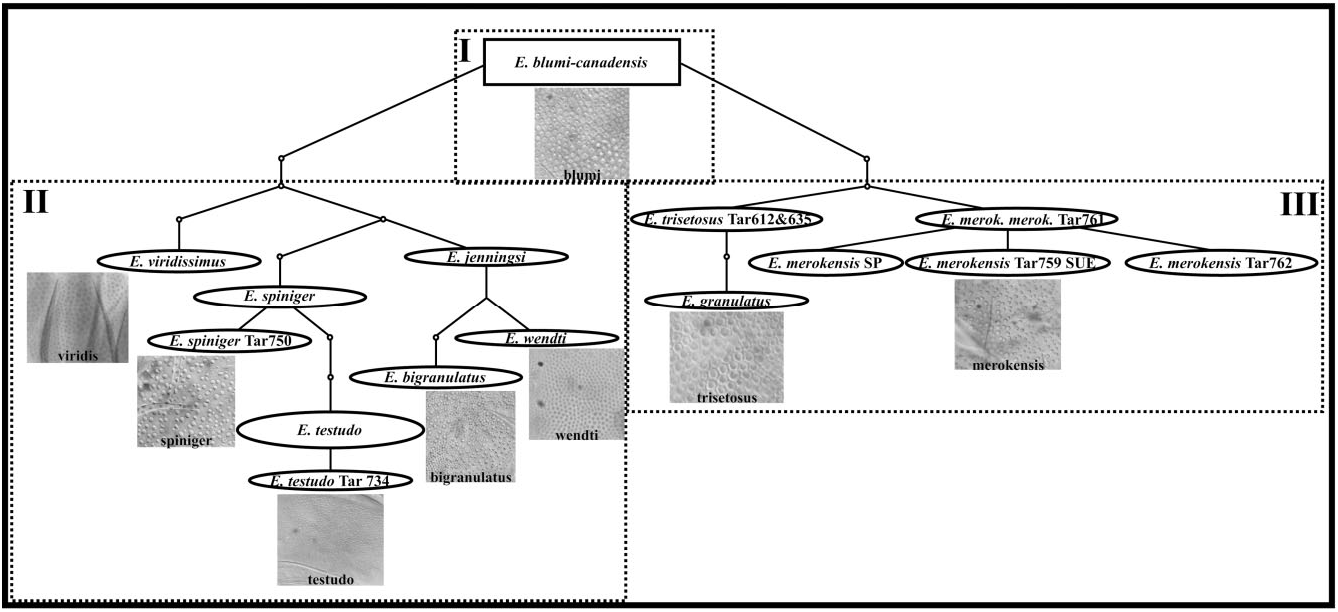
Contrary to the conflict found among the dorsal plate configuration in echiniscid genera, and molecular phylogenetic data (using 18S rRNA and 28S rRNA), cuticular design seems to contain evolutionary signal within Echiniscus . However, the traditional cuticular design groups (see Ramazzotti & Maucci, 1983; Peluffo et al., 2002; Pilato et al., 2007, 2008), i. e. bigranulatus , blumi-canadensis , merokensis , arctomys, and viridis ( Fig. 1 View Figure 1 and Figs S1 View Figure 1 –7), did not coincide with the groups found in our parsimony network ( Fig. 5 View Figure 5 ). The three groups of Echiniscus , supported by the AMOVA (named I, II, and III; Fig. 5 View Figure 5 ), show different types of cuticular design. One group (group I, Fig. 5 View Figure 5 ) is for the E. blumi-canadensis group, with the typical polygonal sculpture of blumicanadensis . Another group (group II) includes species with granulation in their cuticles, from the large, roundish, densely distributed granulation of E. viridissimus to E. spiniger and E. testudo with small roundish pores of different sizes, regularly but not densely distributed, E. jenningsi and E. wendti with similar cuticular designs of very minute granulation regularly and densely distributed ( Dastych, 1984), and finally E. bigranulatus , with mixed large and fine granulation evenly distributed. In the third group (group III), we include two E. blumicanadensis , which could be a case of misidentification ( Guil & Giribet, 2009), E. granulatus (pores structured in polygonal areas; Ramazzotti & Maucci, 1983) and E. merokensis , which shows a cuticle with pores of various sizes and shapes and smooth cuticle between pores ( Fig. 5 View Figure 5 ). In contrast to the evolutionary signal in cuticular design, we found little biogeographical signal in our data even at broad geographical scale: for example, E. blumi from Chile and Greenland share 18S rRNA and/or 28S rRNA haplotypes; this is also the case of E. merokensis from Spain and Greenland, and of E. testudo from Greece, France, and Denmark. This may support the idea that microscopic animals can achieve broad distributions mediated by long-distance passive dispersal ( Fenchel & Finlay, 2004).
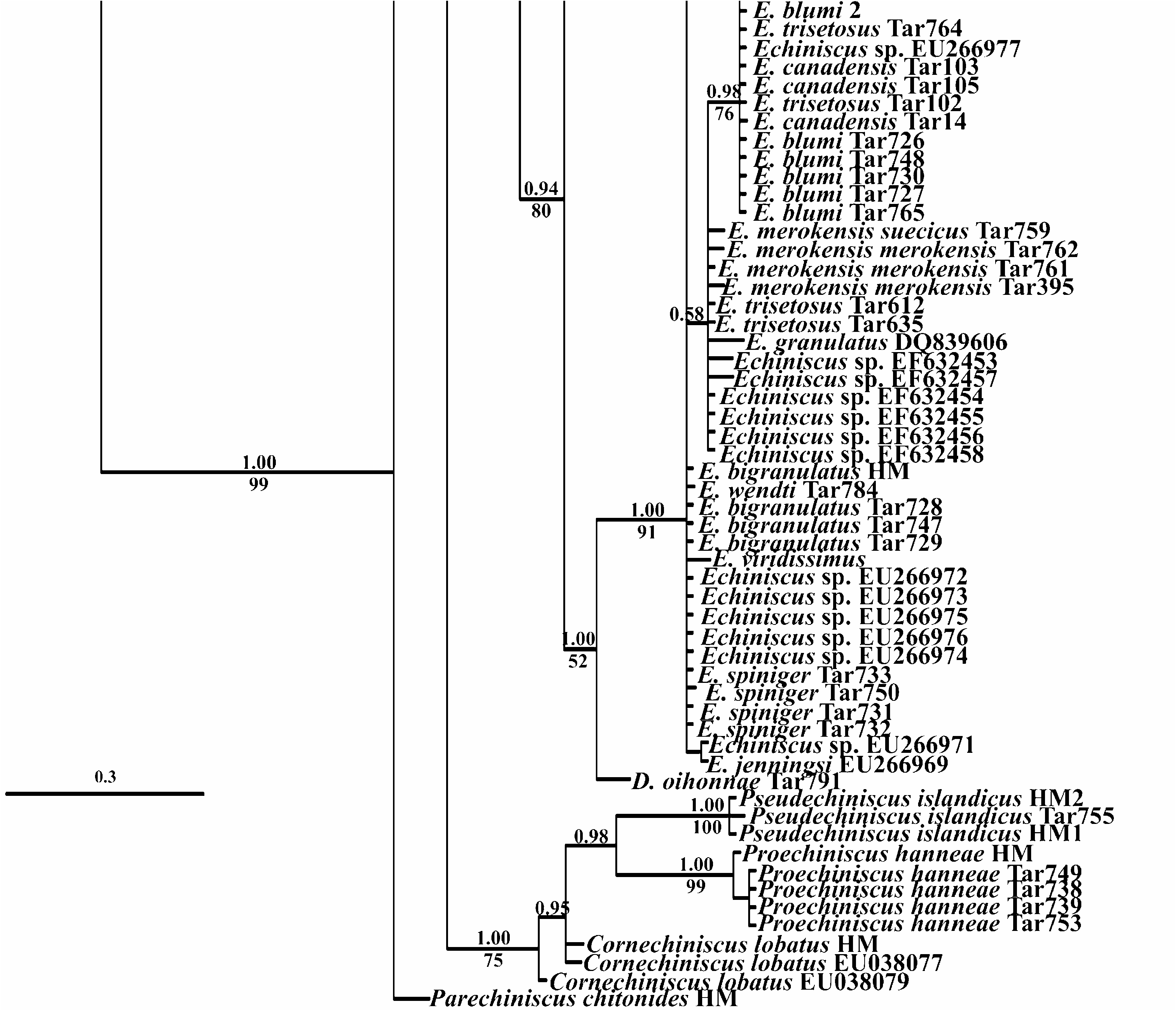
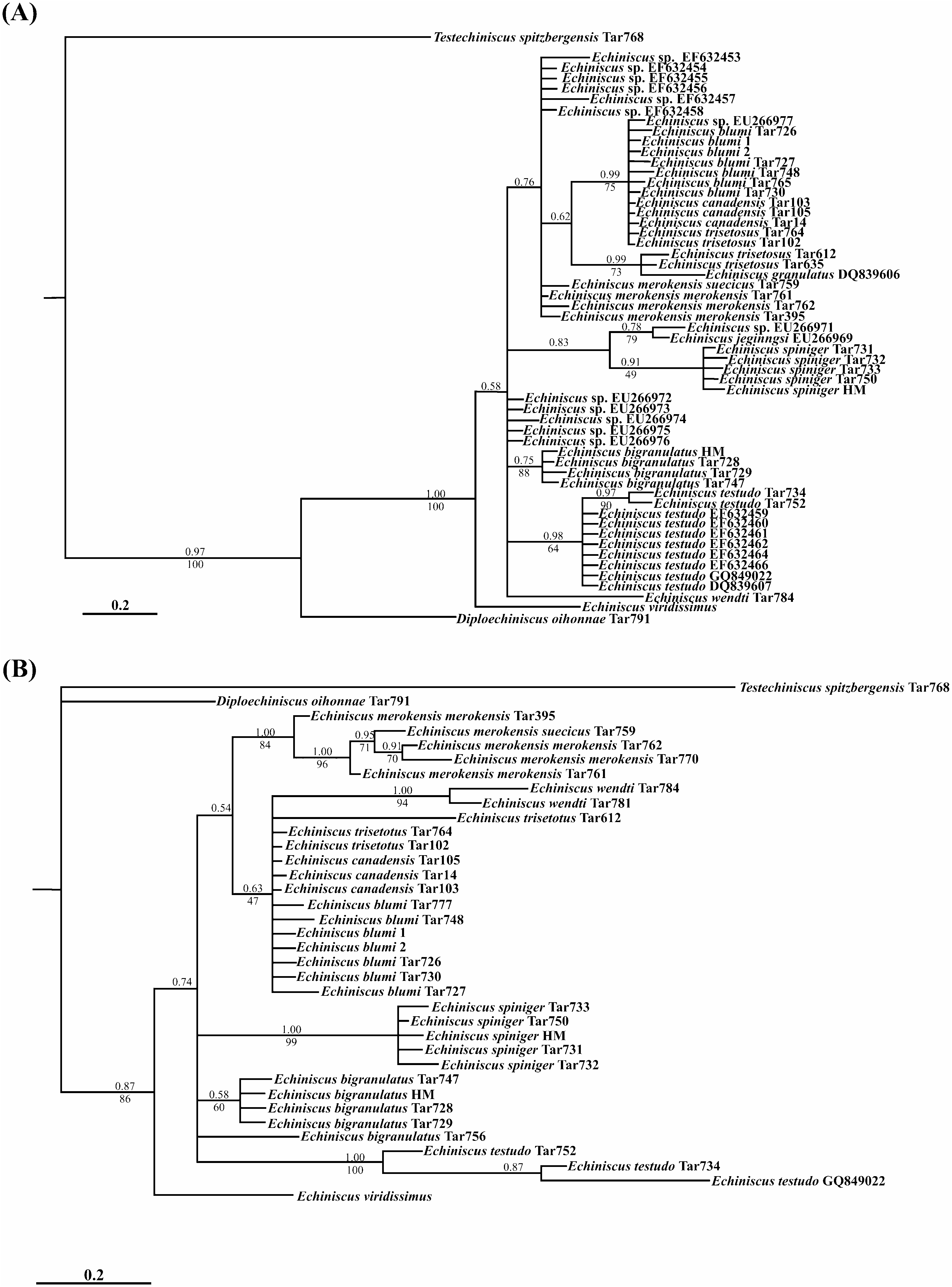
Monophyly of the different Echiniscus species analysed is phylogenetically supported ( Fig. 4 View Figure 4 ), but with different data sets, depending on the species: 28S rRNA supports monophyly of four Echiniscus species ( E. merokensis , E. wendti , E. spiniger , and E. testudo ) while 18S rRNA confirms monophyly of three species ( E. granulatus , E. spiniger , and E. testudo ), and one complex of species ( Echiniscus blumi-canadensis ). Two species or complexes of species remain problematic. Echiniscus bigranulatus is monophyletic except for specimen Tar756 (GenBank accession number: JX114856 View Materials ; Fig. 3 View Figure 3 and Fig. S1 View Figure 1 ). The Echiniscus blumi-canadensis complex (comprising E. blumi , E. canadensis , E. mediantus Marcus, 1930 , E. trisetosus , E. dearmatus Bartoš, 1935 , and probably E. marleyi Li, 2007 ) has not been supported by 28S rRNA data, as opposed to the rest of the Echiniscus species complexes studied ( Fig. 4B View Figure 4 ). In contrast, the blumi-canadensis complex finds 18S rRNA support ( Fig. 4A View Figure 4 ). However, this complex of species has been problematic for a long time, due to high morphological variability ( Guil, 2008) not reflected in the molecular information (at least for COI data; Guil & Giribet, 2009), and apparently supported by the present study (morphospecies of the complex, E. blumi , E. canadensis , and E. trisetosus are not phylogenetically differentiated either by 18S rRNA data or 28S rRNA). Two specimens of the blumi-canadensis complex had different sequences (coded as Tar612 and Tar635; GenBank accession numbers: FJ435717 View Materials , FJ435782 View Materials , FJ435718 View Materials , and FJ435783 View Materials ; Table 1) when compared with the rest of the blumi-canadensis individuals. These two specimens (Tar612 and Tar635) were closely related to E. granulatus , indicating a possible misidentification (for these GenBank sequences there is no morphological voucher).
With the current sampling two clear trends are noted: (1) the distribution of plates within the family Echiniscidae is in conflict with the phylogenetic information derived from 18S and 28S rRNA sequence data; and (2) the cuticular design contains evolutionary signal congruent with the 18S rRNA information within Echiniscus . Together with morphological and any other source of information, this would contribute towards a more integrative taxonomic approach within this group of minute animals. We also emphasize the importance of generating and making available morphological information for the study of these tiny animals, as argued previously by Pleijel et al. (2008).
No known copyright restrictions apply. See Agosti, D., Egloff, W., 2009. Taxonomic information exchange and copyright: the Plazi approach. BMC Research Notes 2009, 2:53 for further explanation.