Mesobiotus philippinicus, Mapalo, Marc A., Stec, Daniel, Mirano-Bascos, Denise & Michalczyk, Łukasz, 2016
|
publication ID |
https://doi.org/10.11646/zootaxa.4126.3.6 |
|
publication LSID |
lsid:zoobank.org:pub:3DA855B5-CA5A-4ACC-AE92-162472A6F7E2 |
|
DOI |
https://doi.org/10.5281/zenodo.6081837 |
|
persistent identifier |
https://treatment.plazi.org/id/7F0987A6-FF80-FFDC-ACC6-F905FB96F875 |
|
treatment provided by |
Plazi |
|
scientific name |
Mesobiotus philippinicus |
| status |
sp. nov. |
Mesobiotus philippinicus View in CoL sp. nov.
( Tables 4 View TABLE 4 –6, Figs 1–51 View FIGURES 1 – 5 View FIGURES 6 – 9 View FIGURES 10 – 16 View FIGURES 17 – 20 View FIGURES 21 – 36 View FIGURES 37 – 51 )
Description of the new species. Animals (measurements and statistics in Table 4 View TABLE 4 ).
Body white/transparent ( Figs 1–2 View FIGURES 1 – 5 ), eyes present (also in mounted individuals). Under PCM, cuticle appears completely smooth with no granulation visible (except in some specimens a very fine granulation barely detectable on legs IV). Under SEM, however, extremely fine granulation (microgranules, ca. 0.05 µm in diameter) is visible on the entire dorso-lateral cuticle ( Figs 3–5 View FIGURES 1 – 5 ). The microgranulation is scattered evenly on most of the cuticle, but is denser to the caudo-dorsal region. Moreover, patches of dense, raised granule aggregates are present on external cuticle of all legs, above claws ( Figs 6–9 View FIGURES 6 – 9 ). These raised granule aggregates are ca. 0.2 µm in diameter, and consist of two to over thirty microgranules, but more often between five and ten microgranules ( Figs 7, 9 View FIGURES 6 – 9 ). Patches on legs I–III are smaller ( Figs 6–7 View FIGURES 6 – 9 ) than those on legs IV ( Figs 8–9 View FIGURES 6 – 9 ). Cuticular pores absent.
Bucco-pharyngeal apparatus of the Macrobiotus type ( Fig. 10 View FIGURES 10 – 16 ), with ten peribuccal lamellae. Oral cavity armature of the harmsworthi- type, composed of three bands of teeth visible in both PCM and SEM ( Figs 10–16 View FIGURES 10 – 16 ). The first band appears as small granules/cones placed just behind (sometimes also at the base) of the lamellae under PCM/SEM, respectively, and are arranged in 5–7 irregular rows (PCM: Figs 10–14 View FIGURES 10 – 16 ; SEM: 15–16, filled arrowheads). Teeth in the second band ( Figs 15–16 View FIGURES 10 – 16 , empty arrowheads) are intermediate in size between those of the first and third bands of teeth, and are in the shape of small ridges parallel to the longitudinal axis of the buccal tube ( Figs 10–14 View FIGURES 10 – 16 ). The second band is continuous with the teeth arranged in one row and are positioned in the posterior portion of the oral cavity just behind the ring fold ( Figs 19–20 View FIGURES 17 – 20 , rhomboid) and just before the third band of teeth ( Figs 15–16 View FIGURES 10 – 16 , marked “L” and “M”). Situated posteriorly in the oral cavity just behind the second band of teeth and just before the buccal tube opening are the third band of teeth comprising three dorsal, distinctly separated, long and thin ridges ( Fig 15 View FIGURES 10 – 16 “L” and “M”) and three to six ventral teeth: two lateral ridges ( Fig 16 View FIGURES 10 – 16 "L") and 2–4 round or oval median teeth ( Fig 16 View FIGURES 10 – 16 “M1–4”). Under PCM, the latero-ventral teeth are wing-shaped, thicker in their median portions ( Figs 10–14 View FIGURES 10 – 16 ), and under SEM are flat and serrated, with their height almost twice that of the teeth in the second band ( Figs 15–16 View FIGURES 10 – 16 ). Buccal tube rigid and slender with the ventral lamina ( Fig 10 View FIGURES 10 – 16 ). The pharyngeal bulb is equipped with pharyngeal apophyses, three macroplacoids and a microplacoid, all spaced equidistant from each other ( Fig. 10 View FIGURES 10 – 16 and the upper insert). The first and third macroplacoids are rod-shaped whereas the second macroplacoid is granular. The first macroplacoid is thinner anteriorly whereas the third macroplacoid has a sub-terminal constriction. The macroplacoid sequence is 2<3<1. In the majority of individuals, the drop-like microplacoid is larger than the second macroplacoid ( Fig. 10 View FIGURES 10 – 16 , Table 4 View TABLE 4 ).
Claws of the Mesobiotus type, with a septum and well-developed accessory points ( Figs 17–20 View FIGURES 17 – 20 ). Lunules smooth in claws I–III ( Figs 17–18 View FIGURES 17 – 20 ) and lightly to moderately crenulated in claws IV ( Figs 19–20 View FIGURES 17 – 20 ). Lunules, connected to the claw with a peduncle, present in all claws ( Fig. 17–20 View FIGURES 17 – 20 ). Double W-shaped transverse bars under claws I–III ( Fig. 17 View FIGURES 17 – 20 , filled arrowhead); a horseshoe-shaped structure connects anterior and posterior lunules IV ( Fig. 19 View FIGURES 17 – 20 ).
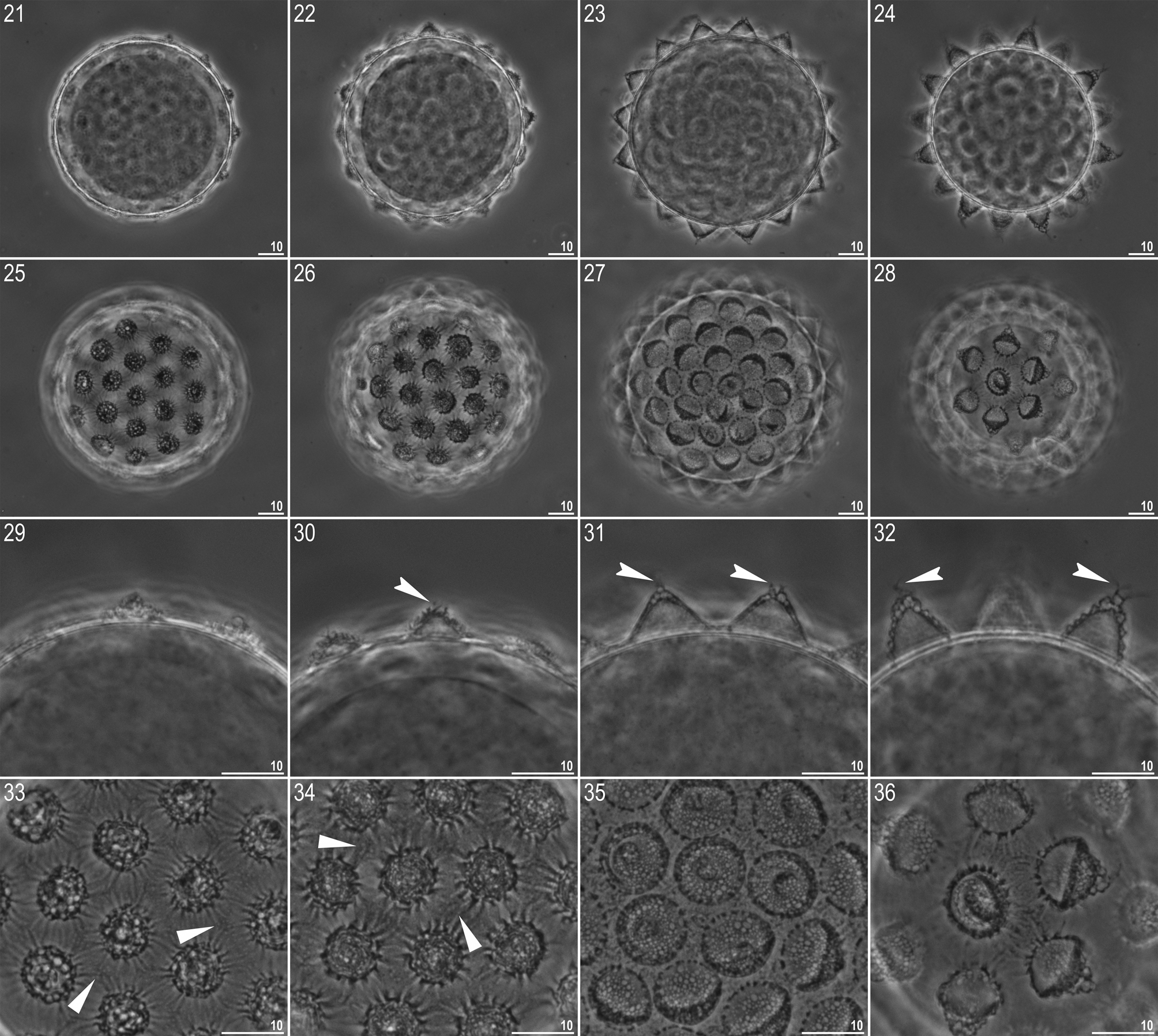
Egg (measurements and statistics in Table 5 View TABLE 5 ). White, laid free, spherical in shape and equipped with conical to dome-shaped processes ( Figs 21–32 View FIGURES 21 – 36 , 37–39 View FIGURES 37 – 51 ). The processes are evenly spaced and exhibit considerable variability in size and shape between eggs: from low domes to high cones ( Figs 21–24, 29–32 View FIGURES 21 – 36 , 37–39, 46–48 View FIGURES 37 – 51 ). Process surface covered with wrinkles forming a rose-like whorl, clearly identifiable under SEM ( Figs 40–48 View FIGURES 37 – 51 ) but under PCM identifiable only as serration on an intersected process wall ( Figs 30, 32 View FIGURES 21 – 36 ). Very rarely processes have smooth walls ( Fig. 31 View FIGURES 21 – 36 ). The labyrinthine layer is visible under PCM as a reticulum in process walls, with mesh size varying considerably between eggs ( Figs 33–35 View FIGURES 21 – 36 ). Some processes are terminated with one to several short but thin and flexible portions, visible in both PCM ( Figs 30–32 View FIGURES 21 – 36 , indented arrowheads) and in SEM ( Figs 49–51 View FIGURES 37 – 51 ), covered irregularly with small granules that are visible only in SEM ( Figs 49–51 View FIGURES 37 – 51 ). Small pores (0.3 µm) are present on the bases of some processes and scattered across the inter-process surface. The pores are clearly visible in SEM ( Figs 40–48 View FIGURES 37 – 51 ) but only occasionally detectable under PCM ( Figs 33–34 View FIGURES 21 – 36 , filled arrowheads). Egg surface clearly wrinkled, with wrinkles radiating out from the process bases, but not forming a connective network ( Figs 33–36 View FIGURES 21 – 36 , 40–48 View FIGURES 37 – 51 ).
DNA sequences. A single haplotype was found for each sequenced marker across the eight analysed individuals, thus only one sequence per locus was deposited in the GenBank.
The 18S rRNA sequence (GenBank: KX129793 View Materials ), 1690 bp long:
TCTAAGTACTTGCTTTAACAAGGCGAAACCGCGAATGGCTCATTAAATCAGTTATGGTTCACTAGATCGTATATCCTACA CGGATAACTGAGGTAATTCTTCAGCTAATACGTGCTACAAGCTCGTTCCCTTGTGGAGCGAGCGCAGTTATTAGAACAAA ACCAATCCGGCCTTCGGGTCGGTTAAAATGGTGACTCTGAATAACCGAAGCGGAGCGTATGGTCTCGTACCGACGCCAGA TCTTTCAAGTGACTGCTCTATCAGCTTGTTGTTAGGTTATGCTCCTAACAAGGCTTCAACGGGTAACGGGGTATCAGGGT CCGATACCGGAGAGGGAGCCTGAGAAACGGCTACCACATCCAAGGAAGGCAGCAGGCGCGCAAATTACCCACTCCCAGCA CGGGGAGGTAGTGACGAAAAATAACGATGCGAGGGCATATTGCTTCTCGTAATCGGAATGGGTACACTTTAAATCCTTTA ACGAGGATCTATTGGAGGGCAAGTCTGGTGCCAGCAGCCGCGGTAATTCCAGCTCCAATAGCGTATATTAAAGTTGCTGC GGTTAAAAAGCTCGTAGTTGATCGTAGACGTAGGATGAGTGGTGCGCTTTCCGGCGCTACTGCTTGTTCCGACGTCAAAA GCCGGTCATGTCTTGCATATCCTTCACTGGGTGTGCTTGGCGGCCGGAGCGTTTACTTTGAAAAAATTAGAGTGCTCAAA GCAGGCGTACGGCCTTGCATAATGGTGCATGGAATAATGGAATAGGACCTCGGTTCTATTTTGTTGGTTATCGGAGCTCG AGGTAATGATTAAGAGGAACAGACGGGGGCATTCGTATTGCGGCGTTAGAGGTGAAATTCTTGGATCGTCGCAAGACGCA CTACTGCGAAAGCATTTGCCAAGAATGTTTTCATTAATCAAGAACGAAAGTTAGAGGTTCGAAGGCGATCAGATACCGCC CTAGTTCTAACCATAAACGATGCCAACCAGCGATCCGCCGATGTTCTTTTGATGACTCGGCGGGCAGCTTCCGGGAAACC ACAAGTGCTTAGGTTCCGGGGGAAGTATGGTTGCAAAGCTGAAACTTAAAGGAATTGACGGAAGGGCACCACCAGGAGTG GAGCCTGCGGCTTAATTTGACTCAACACGGGAAAACTTACCCGGCCCGGACACTGTAAGGATTGACAGATTGAGAGCTCT TTCTTGATTCGGTGGGTGGTGGTGCATGGCCGTTCTTAGTTGGTGGAGCGATTTGTCTGGTTAATTCCGATAACGAACGA GACTCTAGCCTGCTAAATAGCCAACCGATCCGCAGCGTCGGTTGCTACAAAAGCTTCTTAGAGGGACAAGTGGCGTTTAG TCACATGAGATTGAGCAATAACAGGTCTGTGATGCCCTTAGATGTCCGGGGCCGCACGCGCGCTACACTGAAGGGATCAG CGTGCTTAATCGCCTTGCCCGGAAGGGCTGGGGAATCCGATTAAACCCCTTCGTGATTGGGATTGAGCTTTGTAATTATC GCTCATGAACGAGGAATTCCCAGTAGGCACGAGTCATAAGCTCGTGTCGATTAAGTCCCTGCCCTTTGTACACACCGCCC GTCGCTACTACCGATTGAACCCTTTAGTGAGGTCTTCAGACCGGCCGTGGCGGCAGACTTTGTCTGCAGCTCACAGGTTG GGAAGACGAC
The 28S rRNA sequence (GenBank: KX129794 View Materials ), 755 bp long:
CTGCGAGTGAAACGGGACCAGCCCATCGCCGAATCCTTTCTGGCAACAGTGAAGGAACTGTGGCGTGTAGAAGATGTCTG CCGGTGTGGTTAGTTTGCGTAAGTTCTCCTGAGTGAGACTCCATCCCATGGAGGGTGCAAGGCCCGTACCGCAAGCAGCT GATGCCGGTTAGCGTCTTTCGGAGAGTCGCCTTATTTGTGAGTATAAGGTGAAGTCGGTGGTAAACTCCATCGAAGGCTA AATATGGCCGCGCGTCCGATAGCGAACAAGTACCGTGAGGGAAAATTGAAAAGCACTTTGAAGAGAGAGCGAAACAGTGC GTGAAACGGCTCAGAGGCAAGCAGATGGGGCCTCGAAGGCAGAGCAGCGAATTCAGCCGGTAGTTTGTGCGGATGGCGGT TGTCGGGATCGTAAGACTCGGTGGCTGTCAGTGCTGTGTGTACAACTGCTGGTGCACTTTCGTTGCACGATACACAACCG CCGTTGAGCGAGCATCCGTTGAGCTGGCAGCGTGGAGCCTTATTCCCCTTAAAAAGGCATAGGTGCTTACAGCTGGCTTT AACGCGTTTGCGCCTCAACCGGTCAAGTCTATGTATGCCATGCTTCGGCAGTGGGACCGGCTTGCTCCGATTTGGCGTTA GGAATGACGACTTTGTCGGCTTCTTGGCGTTAGGTGGACTCGATGTCGGTTATCCACGTAGGCATATTGTTGATTCGGTT GTGAGTAGATGGCAGCCTATCTAACCCGTCTTGAA
The ITS-2 sequence (GenBank: KX129795 View Materials ), 481 bp long:
AGAACGCAGCAAGATGTGAGACGTAACTGGAATTGCAGGACTTGTGACAGTTTATTCTTCGAATGCAAATCGCGGCGTTG GGTTAACCGACGCCACGCCTGGTTGAGGGTCAGTTGAATTAAACAAATCATAGTCGCGCAATGATTATGGATTGTCTGAA CAGTGCGTCAAGCACTGGCAGATGAAGTACAGACCCCAGACGTTGAGTGTCATGCGTACCGCTGCTGGTGCGATTGACGT TATATGGGACTTGTGCCGCAGTTGCACAGTAAACAGTCTCATGCACGTCCGATCGGCAAGTCGGCCTCGCTGCACAACCG CTGTTAGCTATAGCGGTAAGCGATATAGCGTCTGTGTATAAAACGACGTTTGACAACAAGCGATCGATCGGTCAATTGCT CGACTAATCGTCGCACAATCGTCGGATACTTTTTCTTTTGACCTCAGCTCAGACGAGATTACCCGCTGAACTTAAGCATA T
The COI sequence (GenBank: KX129796 View Materials ), 763 bp long:
TTTTTTTTTGGATTATGAGCAGCTACGGTAGGTACATCACTTAGAATAATCATTCGATTAGAGCTAAGACAACCTGGAAG ATTATTTAATGACGAACAATTCTATAATGTAGTTGTAACTAGACACGCATTTATTATAATTTTTTTTTTTGTTATACCTA TTCTGATCGGTGGATTTGGTAACTGACTAGTTCCACTTATACTAAATGCACCTGACATAGCATTTCCCCGAATAAATAAC CTAAGATTTTGATTACTTCCTCCATCTTTAATTTTAATTTTATCTAGTTCAATCGTAGAACAGGGTGCAGGCACCGGATG AACTGTTTATCCGCCCCTATCAAGATTTTTTGCGCATAGAGGACCTAGAGTCGACATAACCATTTTTTCCCTTCATGTAG CTGGAGCTTCCTCAATTCTTGGGGCTATCAACTTTATTACAACTATTTTAAACATACGAATACATAATATGACAATAGAA CTACTACCCCTATTTGTATGATCAGTTCTTCTTACTGCTATTTTATTGCTCTTATCCCTCCCTGTACTTGCAGGTGCAAT CACTATATTACTTTTAGATCGAAATTTTAACACCTCCTTTTTCGACCCAAGAGGAGGAGGAGACCCTATTCTATACCAAC ACTTATTTTGATTCTTCGGTCATCCCGAAGTCTATATTTTAATTCTTCCTGGGTTTGGAATTATTTCACAACTAATTATC CATTTCAGTGGAAAACATTTAACTTTCGGGCATATCGGAATGA
Type locality. Locale: 14°38'54.4"N, 121°04'14.8"E, 62 m a.s.l.: UP Science Park, near the Institute of Mathematics, University of the Philippines, Diliman, Quezon City, Philippines. Habitat: shady urban park. Substrate: moss on a tree trunk. Coll. Marc Mapalo, Maynard Gian Valdez, Rafael Fernando and Gamaliel Cabria.
Etymology. The new species is named after the Philippines, where it was discovered and documented as the very first eutardigrade reported in the country.
Type depositories. The holotype (slide PH.001.19), 15 paratypes and 18 eggs (slides PH.001.4–7, 10–11, 15– 16, 20–22) are preserved at the Protein Structure and Immunology Laboratory of the National Institute of Molecular Biology and Biotechnology, University of the Philippines-Diliman, Quezon City, Philippines; 16 paratypes, 14 eggs and 15 chorions (slides PH.001.1–3, 8–9, 12–14, 17–18, 23–24) are preserved at the Department of Entomology, Institute of Zoology, Jagiellonian University, Kraków, Poland.
TABLE 4. Measurements (in µm) of selected morphological structures of individuals of Mesobiotus philippinicus sp. nov. mounted in Hoyer’s medium (N—number of specimens / structures measured. RANGE refers to the smallest and the largest structure among all measured specimens; SD—standard deviation).
| CHARACTER | N | RANGE | MEAN | SD | Holotype |
|---|---|---|---|---|---|
| µm pt | µm pt | µm pt | µm pt | ||
| Body length | 23 | 238 – 442 765 – 1102 | 379 941 | 53 93 | 432 1075 |
| Buccopharyngeal tube | |||||
| Buccal tube length | 24 | 31.1 – 48.1 – | 40.4 – | 4.1 – | 40.2 – |
| Stylet support insertion point | 24 | 24.6 – 37.1 76.4 – 79.8 | 31.6 78.1 | 3.2 1.0 | 31.6 78.6 |
| Buccal tube external width | 24 | 4.5 – 8.0 1 1.8 – 1 7.3 | 6.5 16.0 | 0.8 1.1 | 6.5 16.2 |
| Buccal tube internal width | 24 | 3.7 – 6.1 1 1.0 – 1 6.2 | 5.0 12.4 | 0.6 1.0 | 4.9 12.2 |
| Ventral lamina length | 21 | 18.2 – 26.3 51.4 – 58.8 | 22.2 55.5 | 2.0 2.1 | 22.1 55.0 |
| Placoid lengths | |||||
| Macroplacoid 1 | 24 | 3.9 – 6.9 1 1.3 – 1 4.7 | 5.3 13.0 | 0.7 1.0 | 5.9 14.7 |
| Macroplacoid 2 | 24 | 2.8 – 5.2 7.3 – 11.0 | 3.7 9.1 | 0.6 0.8 | 3.7 9.2 |
| Macroplacoid 3 | 24 | 3.5 – 5.7 10.4 – 13.5 | 4.7 11.6 | 0.6 0.8 | 4.7 11.7 |
| Microplacoid | 24 | 3.1 – 5.7 9.1 – 1 2.3 | 4.3 10.5 | 0.6 0.9 | 4.1 10.2 |
| Macroplacoid row | 24 | 12.7 – 20.5 39.7 – 47.4 | 17.0 42.0 | 1.9 1.8 | 17.1 42.5 |
| Placoid row | 24 | 16.8 – 28.7 50.8 – 60.7 | 22.3 55.2 | 2.7 2.6 | 23.5 58.5 |
| Claw I lengths | |||||
| External primary branch | 20 | 9.3 – 14.1 25.4 – 31.2 | 11.6 28.9 | 1.3 1.4 | 11.4 28.4 |
| External secondary branch | 18 | 7.9 – 12.1 19.5 – 26.3 | 9.7 24.2 | 1.2 1.9 | 8.8 21.9 |
| Internal primary branch | 16 | 8.9 – 13.3 25.7 – 29.7 | 11.0 27.5 | 1.2 1.2 | 10.8 26.9 |
| Internal secondary branch | 13 | 7.4 – 11.9 18.1 – 25.4 | 9.1 22.9 | 1.3 2.0 | 8.9 22.1 |
| Claw II lengths | |||||
| External primary branch | 20 | 11.1 – 14.8 29.3 – 33.5 | 12.8 31.1 | 1.1 1.1 | 13.2 32.8 |
| External secondary branch | 17 | 8.3 – 12.5 21.8 – 28.1 | 10.1 24.9 | 1.1 1.7 | 10.5 26.1 |
| Internal primary branch | 22 | 9.3 – 13.3 24.8 – 30.1 | 11.2 27.6 | 1.2 1.5 | 10.4 25.9 |
| Internal secondary branch | 18 | 6.9 – 10.9 19.7 – 26.7 | 9.4 23.0 | 1.2 2.1 | 9.8 24.4 |
| Claw III lengths | |||||
| External primary branch | 19 | 10.0 – 16.1 27.0 – 34.0 | 12.7 31.3 | 1.5 1.9 | 11.6 28.9 |
| External secondary branch | 14 | 8.9 – 12.5 22.0 – 28.4 | 10.4 25.5 | 1.1 1.8 | 9.2 22.9 |
| Internal primary branch | 16 | 8.8 – 13.2 24.2 – 29.7 | 11.0 27.2 | 1.2 1.4 | 11.0 27.4 |
| Internal secondary branch | 13 | 7.0 – 11.4 19.2 – 26.3 | 9.2 23.6 | 1.3 2.2 | 10.2 25.4 |
| Claw IV lengths | |||||
| Anterior primary branch | 16 | 11.1 – 16.3 29.9 – 35.2 | 13.4 32.6 | 1.5 1.4 | 13.3 33.1 |
| Anterior secondary branch | 16 | 8.6 – 13.2 24.7 – 28.6 | 11.1 27.0 | 1.2 1.1 | 11.5 28.6 |
| Posterior primary branch | 18 | 11.7 – 17.5 33.1 – 38.7 | 14.7 35.7 | 1.5 1.8 | 15.5 38.6 |
| Posterior secondary branch | 17 | 9.7 – 14.5 24.1 – 33.4 | 11.7 28.2 | 1.3 2.2 | 11.0 27.4 |
TABLE 5. Measurements (in µm) of selected morphological structures of eggs of Mesobiotus philippinicus sp. nov. mounted in Hoyer’s medium (N—number of eggs / structures measured, RANGE refers to the smallest and the largest structure among all measured specimens; SD—standard deviation).
| CHARACTER | N | RANGE | MEAN SD |
|---|---|---|---|
| Egg bare diameter | 23 | 62.7 – 79.9 | 70.7 4.5 |
| Egg full diameter | 23 | 71.4 – 97.5 | 83.4 6.6 |
| Process height | 69 | 2.1 – 13.7 | 7.2 2.8 |
| Process base width | 69 | 7.1 – 13.0 | 9.6 1.4 |
| Process base/height ratio | 69 | 76% – 433% | 159% 79% |
| Distance between processes | 69 | 2.4 – 6.7 | 4.6 1.0 |
| Number of processes on the egg circumference | 22 | 15 – 17 | 16.0 0.7 |
No known copyright restrictions apply. See Agosti, D., Egloff, W., 2009. Taxonomic information exchange and copyright: the Plazi approach. BMC Research Notes 2009, 2:53 for further explanation.
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |