Catharanthus
|
publication ID |
https://doi.org/10.1016/j.phytochem.2020.112626 |
|
DOI |
https://doi.org/10.5281/zenodo.8302368 |
|
persistent identifier |
https://treatment.plazi.org/id/8D608791-DF61-1361-FFCD-2944FF78B2C5 |
|
treatment provided by |
Felipe |
|
scientific name |
Catharanthus |
| status |
|
2.8. Molecular modeling of Catharanthus View in CoL View at ENA and Camptotheca P450s
Molecular models for Catharanthus CYP 72A1 and Camptotheca
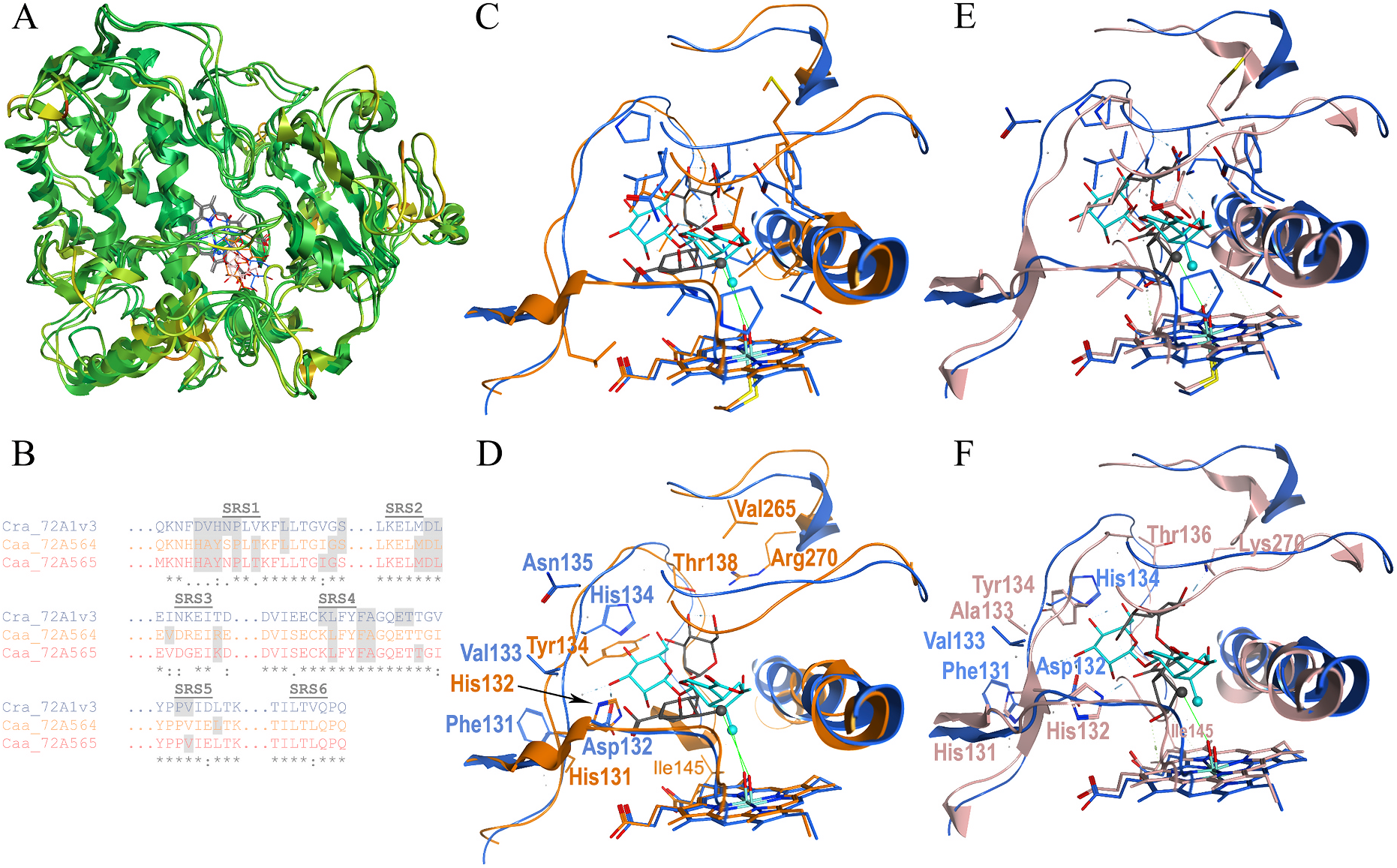
CYP72A564 and CYP72A565, constructed according to published procedures ( Rupasinghe and Schuler, 2006), indicated that the backbones of these three proteins overlay well (RMSD less than 2.0 Å, green, Fig. 7A View Fig ) except for the following regions: the C-terminus of the A ′ -A loop, β1-2, the N-terminus of the C helix, the D-E ′ loop, the N-terminus of the G-helix, the N-terminal half of the H–I loop, the middle of the loop between β1-3 and β1-4, and in a loop region immediately C-terminal of the PERF domain. None of these regions have RMSD values greater than 3.5 Å (orange), and all are outside the predicted SRSs.
Dockings of loganin in the Catharanthus CYP 72A1 catalytic site and loganic acid in the Camptotheca CYP 72A564 and CYP72A565 catalytic sites predicted that both substrates position C10 within 4.5 Å of the oxygen bound to the heme with comparable interaction energies ( Table 3 View Table 3 ); as both proposed mechanisms for the C–C bond scission demand ( Guengerich and Yoshimoto, 2018; Yamamoto et al., 2000), abstraction of a hydrogen from this carbon forms the radical needed to cleave the C–C bond in loganin. Even so, the orientations of these substrates were different with loganic acid oriented more vertically in the two Camptotheca CYP 72A proteins than loganin is in the Catharanthus CYP 72A1 model.
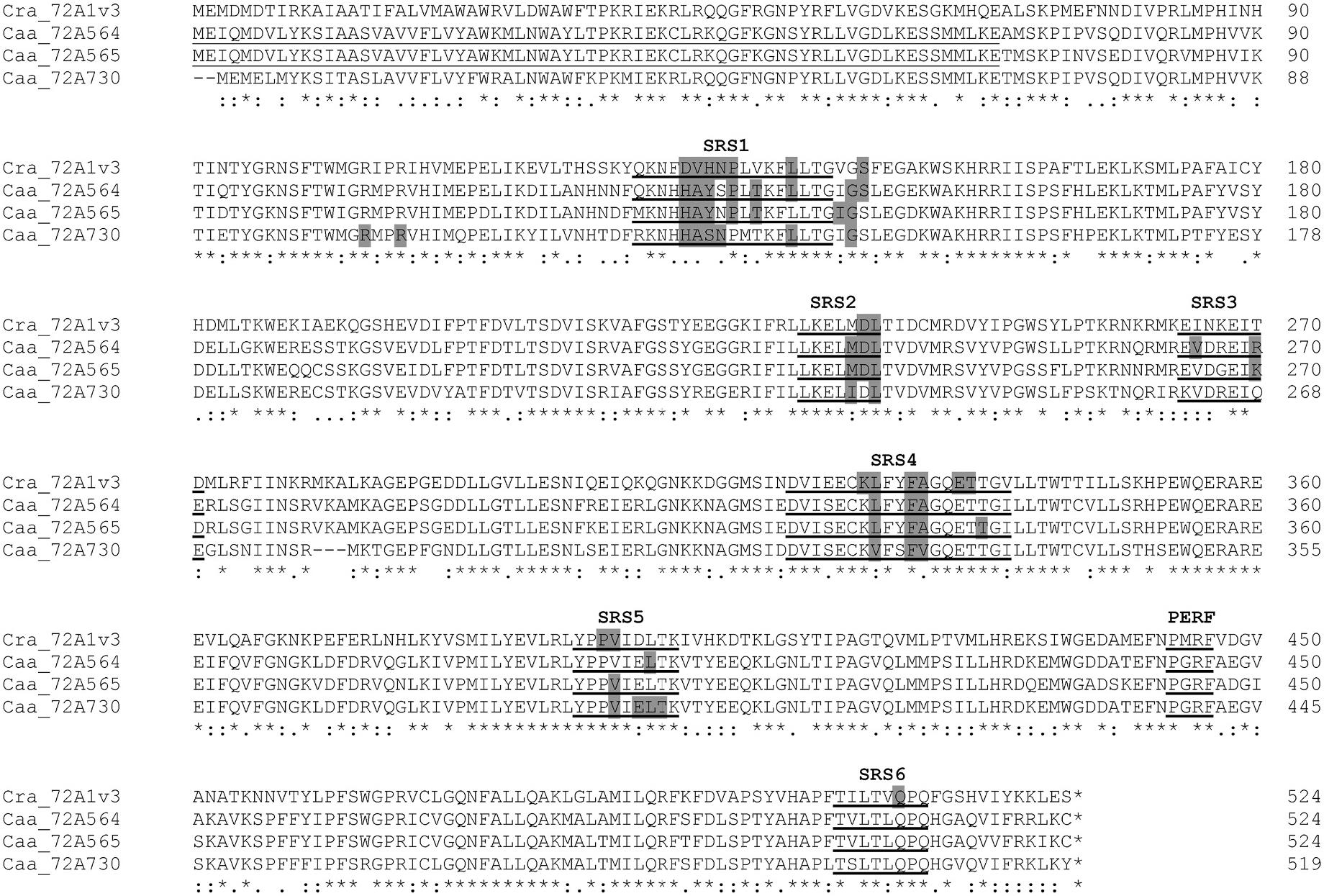
Overlays of the predicted contact residues highlighting amino acids identical in each ( Fig. 7C and E View Fig ) and different in each ( Fig. 7D and F View Fig ) show many identical residues within substrate contact distance and relatively few different residues in this area. Among the variant residues, several differences occur in SRS1 at positions 132, 133, 134, 135 and 138 ( Fig. 2 View Fig ; Fig. 7B, C, 7E View Fig ). The Catharanthus protein differs from the two Camptotheca proteins at all of these positions, and the two Camptotheca proteins differ only at position 135 (Asn/Ser). One additional difference among these three proteins occurs in SRS3 at position 270 with all three proteins having different residues (Thr/Lys/Arg). Similar overlays of identical and different residues in these two Camptotheca proteins ( Fig. 7B–E View Fig ) highlight their substantial identity in all SRSs comprising the catalytic site.
Parameters ± S.E. estimated from a non-linear curve fit of corrected NADPH consumption rates against varying substrate concentrations ( N = 7) from combined technical triplicate assays.
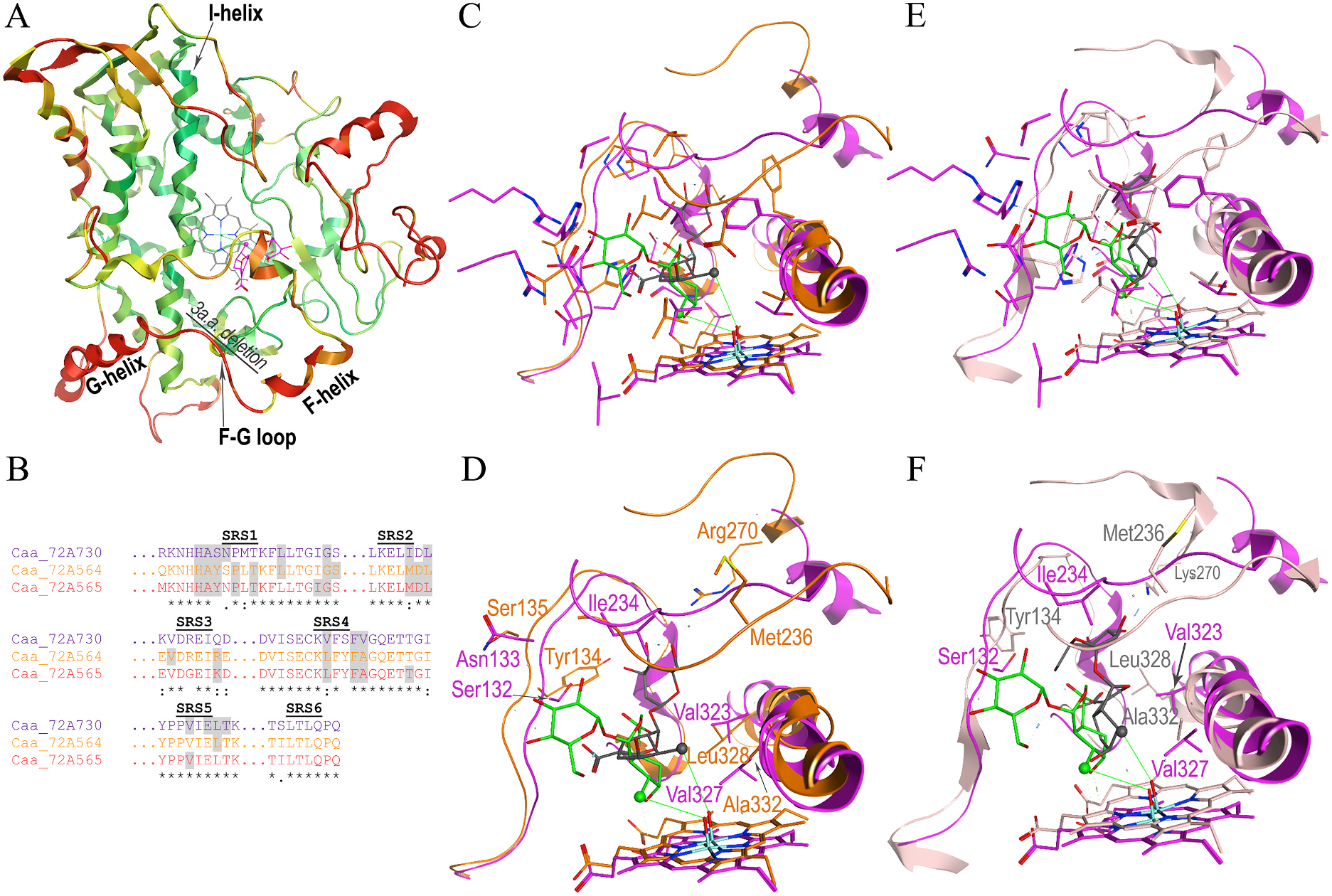
Molecular modeling and docking of Camptotheca CYP 72A730 with loganic acid, which it neither binds nor metabolizes, suggest that this non-substrate can be positioned within the catalytic site ( Fig. 8 View Fig ). The loganic acid in the CYP72A730 model has a similar distance to the oxygen bound to the heme and interaction energy compared to the Camptotheca CYP 72A564 and CYP72A565 models ( Table 3 View Table 3 ). Overlays of the backbones in these three Camptotheca models show the most substantial ( > 4.5 Å) RMSD variance extending from the F-helix through the G-helix (red in Fig. 8A View Fig ) and little in most other regions (≤2.0 Å, green in Fig. 8A View Fig ). Overlays of these three predicted catalytic sites highlighting amino acids identical in each ( Fig. 8C and E View Fig ) and different in each ( Fig. 8D and F View Fig ) show multiple residues that differ within loganic acid contact distance and within the upper regions of its predicted catalytic site. Among the variant residues, several differences occur in SRS1: the Camptoth eca CYP72A730 protein differs from the CYP72A564 and CYP72A565 proteins at positions 132 and 133 (numbered according to CYP72A730) ( Fig. 2 View Fig ; Fig. 8B View Fig ). Differences between these three protein models also occur in SRS2 at position 234, SRS3 at position 268 and SRS4 at positions 323 and 327.
Other differences among these Camptotheca proteins, while occurring in SRS regions ( i.e., SRS4 at position 325, SRS6 at positions 499 and 501) are more than 4.5 Å away from the docked substrate. Notable among the sequence variations within substrate contact distance are those in SRS2 at position 234 and SRS3 at 268 (numbering according to CYP72A730). Because of the substantial RMSD variance in the placement of the F-to G-helices shown in Fig. 8A View Fig , the first of these differences (Ile 234 in CYP72A730), is substantially closer to the docked loganic acid than Met 236 in CYP72A564 and CYP72A565. The second of these (Gln 268 in CYP72A730) is displaced so significantly compared to Arg 270 in CYP72A564 and Lys 270 in CYP72A565 that it is not predicted to lie within substrate contact distance.
The docking poses of their alternate substrate, 7-deoxyloganic acid, in the CYP72A564 and CYP72A565 homology models greatly resembled those of loganic acid in these models (Supplemental Fig. S12). Though considerable variability in the placement of the glucoside moiety is present, the iridoid cores of 7-deoxyloganic acid and loganic acid remain nestled between SRS1 and SRS4. Comparison of the contacts amongst the docked substrate-P450 pairs revealed that the residues predicted as essential for loganic acid docking (especially His132 hydrogen bonding to the carboxylic acid) are likewise predicted contacts for 7-deoxyloganic acid.
No known copyright restrictions apply. See Agosti, D., Egloff, W., 2009. Taxonomic information exchange and copyright: the Plazi approach. BMC Research Notes 2009, 2:53 for further explanation.
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |