Formica nigerrima Nylander 1856
|
publication ID |
https://doi.org/10.11646/zootaxa.5435.1.1 |
|
publication LSID |
lsid:zoobank.org:pub:121D0891-6348-49DB-B96D-7EE0CC6E62D3 |
|
persistent identifier |
https://treatment.plazi.org/id/945A3D69-FF92-FFBF-8394-A8C4FB8CFC30 |
|
treatment provided by |
Plazi |
|
scientific name |
Formica nigerrima Nylander 1856 |
| status |
|
Formica nigerrima Nylander 1856 [ type investigation]
The taxon has been described from a dry place near Montpellier, France (“Locus aridis prope Monspelium”. Investigated was the neotype worker (fixed by Seifert et al. 2016) and additional two workers from the neotype nest, labelled “FRA: 43.6843° N, 3.8763° E, Prades-le-Lez-1.5 SE, 88 m, monodomous colony, Kaufmann 2012.04.30 - K”; depository SMN Görlitz. A wild-card run in a 5-class LDA considering the five morphologically separable entities of the T. nigerrimum species complex allocates the neotype sample with p=0.9954 to the T. nigerrimum cluster.
Material examined. Numeric phenotypical data were taken in 21 samples with 73 workers. They originated from France (18 samples) and Spain (3). For details see supplementary information SI1, SI2 .
Geographic range. So far only known from ten sites within a 20 000 km² area in S France and the eastern Catalan Pyrenees, delimited by 1.78°E and 5.24°E and 42.33°N and 44.03°N. The altitudinal range extends from sea level up to 1561 m in the Catalan Pyrenees GoogleMaps .
Diagnosis:—Worker ( Tab. 3): All shape ratios given below are, in contrast to those in Tab. 3, primary ratios without RAV and all data are given as arithmetic mean ± standard deviation. Largest Palaearctic species, CS 1029 ± 193 µm. Head very broad CL/CW 1.004 ± 0.069. Postocular distance rather small and excavation of hind margin of vertex large, PoOc/CL 0.380 ± 0.010, ExOcc 2.99 ± 1.33 %. Anteromedian clypeal excision rather deep and wide, ExCly/CS 9.85 ± 0.89 %, ExClyW 6.70 ± 0.64 %. The posterior, semicircular end of clypeal excision forms a concave plane delimited by a sharp ventral and a less sharp dorsal edge. Sum of pubescence hairs and smaller setae protruding across the margin of clypeal excision including its dorsal edge very large, nExCly 16.9 ± 6.4. Scape moderately long, SL/CS 0.929 ± 0.052. Minimum distance of the inner margins of antennal socket rings moderately large, dAN/CS 0.302 ± 0.006. Eye smaller than in the supercolonial related species, EL/CS 0.226 ± 0.016. Metanotal groove deep, MGr/CS 4.09 ± 0.76 %. Mesosoma shorter and narrower than in the supercolonial related species, ML/CS 1.222 ± 0.032, MW/CS 0.618 ± 0.012. Second funiculus segment shorter than in the supercolonial related species, Fu2L/CS 13.52 ± 0.45 %, IFu2 1.839 ± 0.091. All body parts including appendages covered by a rather dense pubescence. Setae on dorsal and lateral surfaces of head and mesosoma absent. Longe seta are found on hind margin of 3rd and 4th gaster segment, ventral parts of coxae and anterior clypeus. All body parts blackish brown. Mandibles, edge of clypeus and sometimes antennal funiculus and tarsae with an orange or reddish color component.
Taxonomic comments. The T. nigerrimum group contains five entities separable by NUMOBAT and microsatellite data: the largely monodomous T.nigerrimum and T.hispanicum n. sp. and the supercolonial T. ibericum , T. dario i and T. magnum. Within a total of 129 samples belonging to this group and being both phenotyped and genotyped, the agreement of NUMOBAT and genetic classification was 97.7% which is a really good performance considering the morphological crypsis of the species. Below we describe the run of exploratory and hypothesis-driven data analyses in two blocks.
Block 1: Species hypothesis formation based on NUMOBAT data
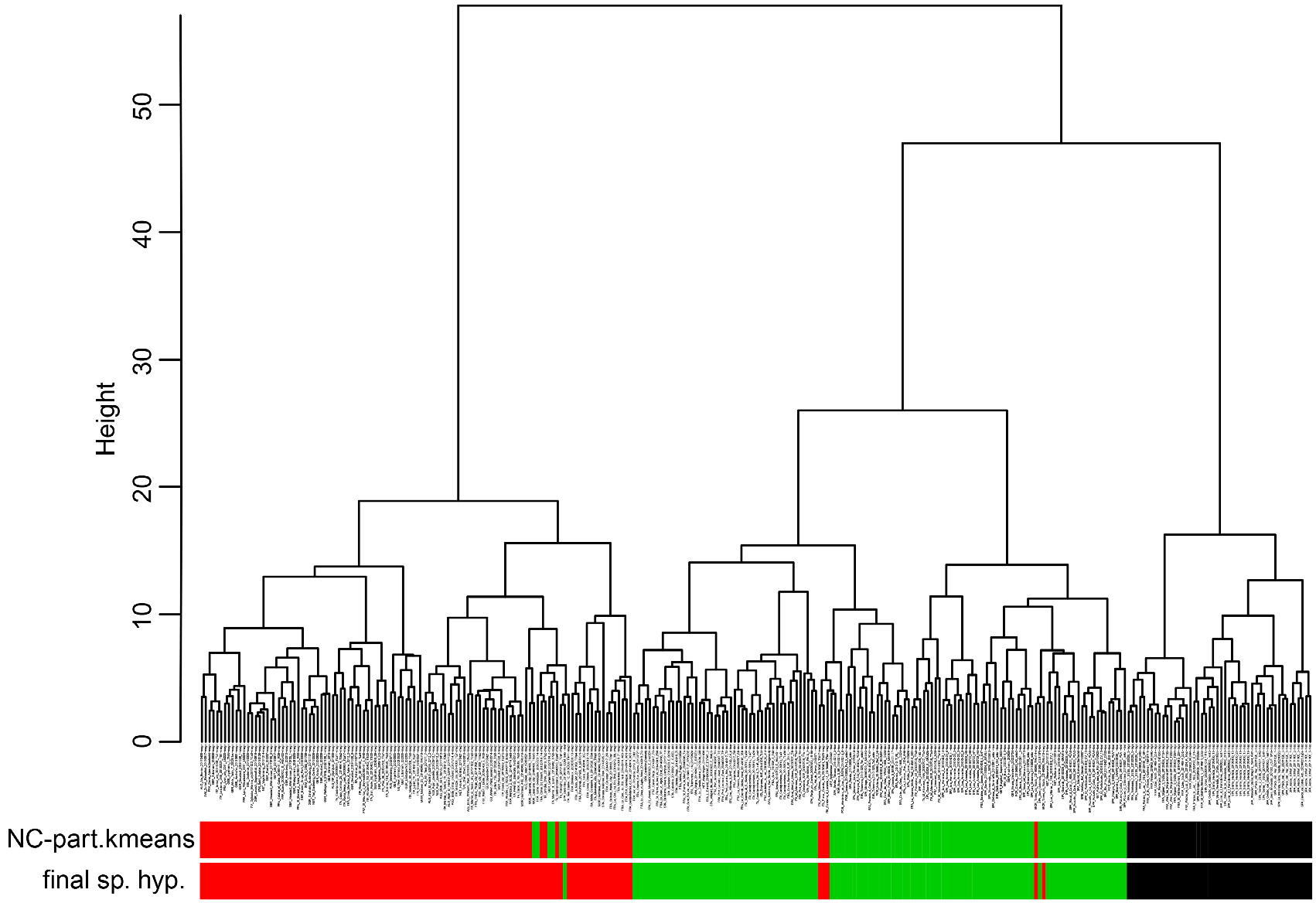
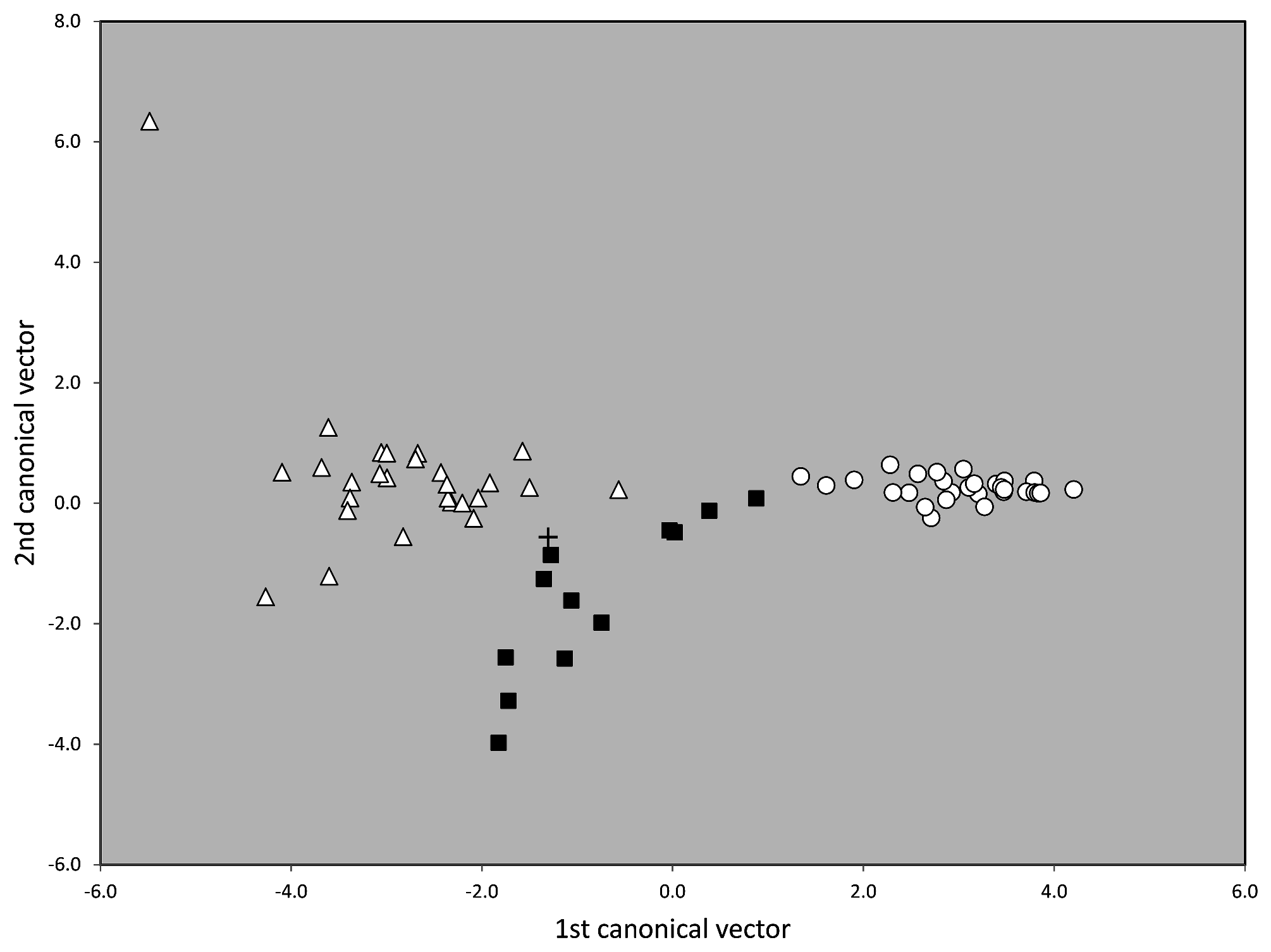
This included in the first step of analysis running all five entities in NC-part.kmeans and NC-Ward using CS and 14 RAV-corrected characters (except PoOc/CL 900 were data were incomplete). Within the 288 nest samples analyzed, the exploratory data analyses revealed three main clusters ( Fig.42 View FIGURE 42 ). The first main cluster (composed of 48 samples of T. nigerrimum and T. hispanicum n. sp.) was demonstrated by the EDAs with 100 % agreement whereas the classification of the second (composed of 124 samples of T. darioi and T. ibericum ) and the third main cluster (composed of 116 samples of T. magnum) disagreed in 2.3 % of the samples.
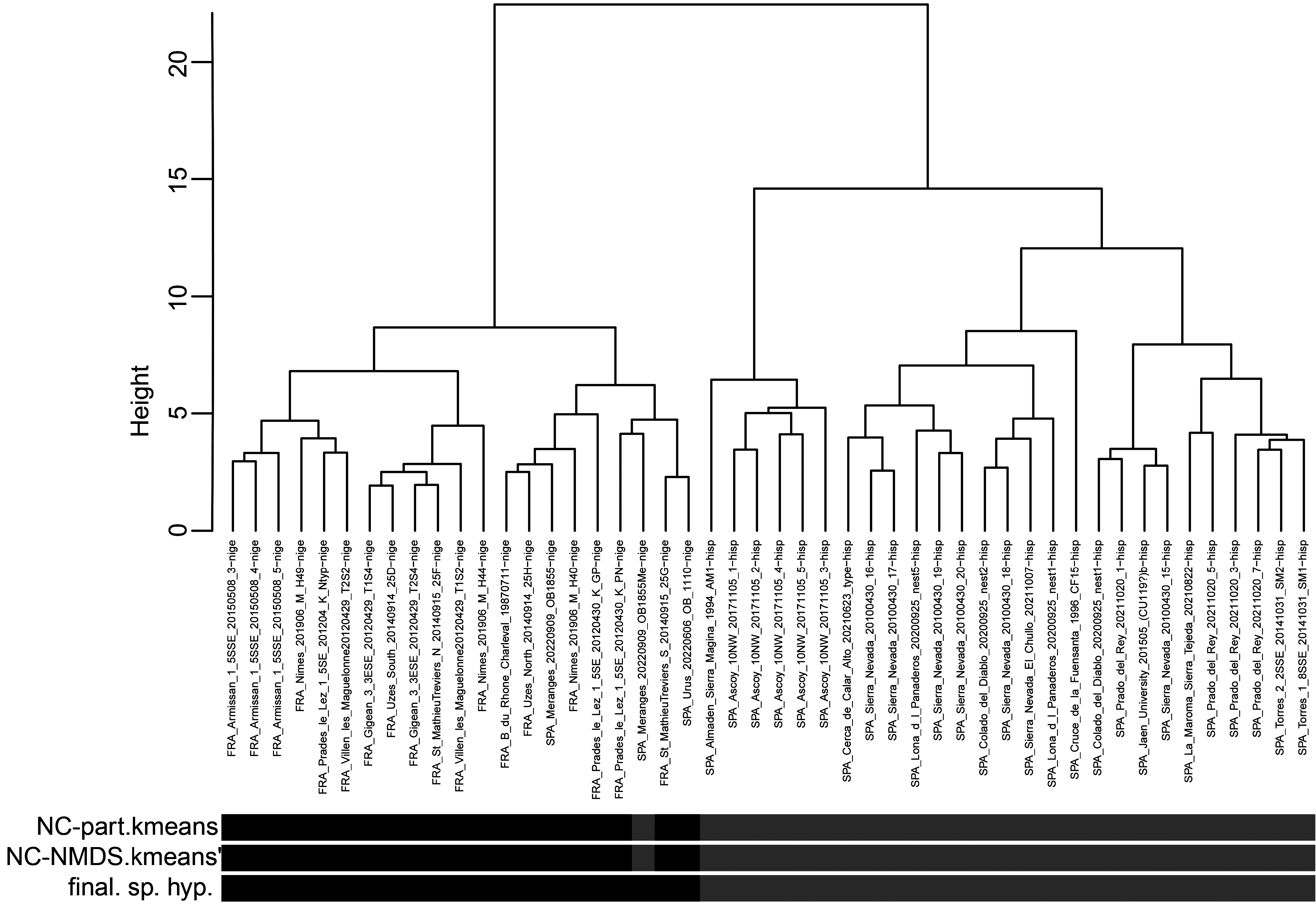
In a second step of analysis the first main cluster, the 48 samples of T. nigerrimum and T. hispanicum n. sp., were run in NC-Ward, NC-part.kmeans and NC-NMDS.kmeans using all 15 characters ( Fig.43 View FIGURE 43 ). Disagreement between the EDAs was given in a single sample the determination of which was rectified as belonging to T. nigerrimum by a wild-card run in a LDA.All nest samples were classified by the LDA with posterior probabilities of p>0.893 and the type series of T. nigerrimum and T. hispanicum n. sp. were allocated in wild-card runs to the corresponding clusters with p = 0.999 and p = 0.998 respectively.
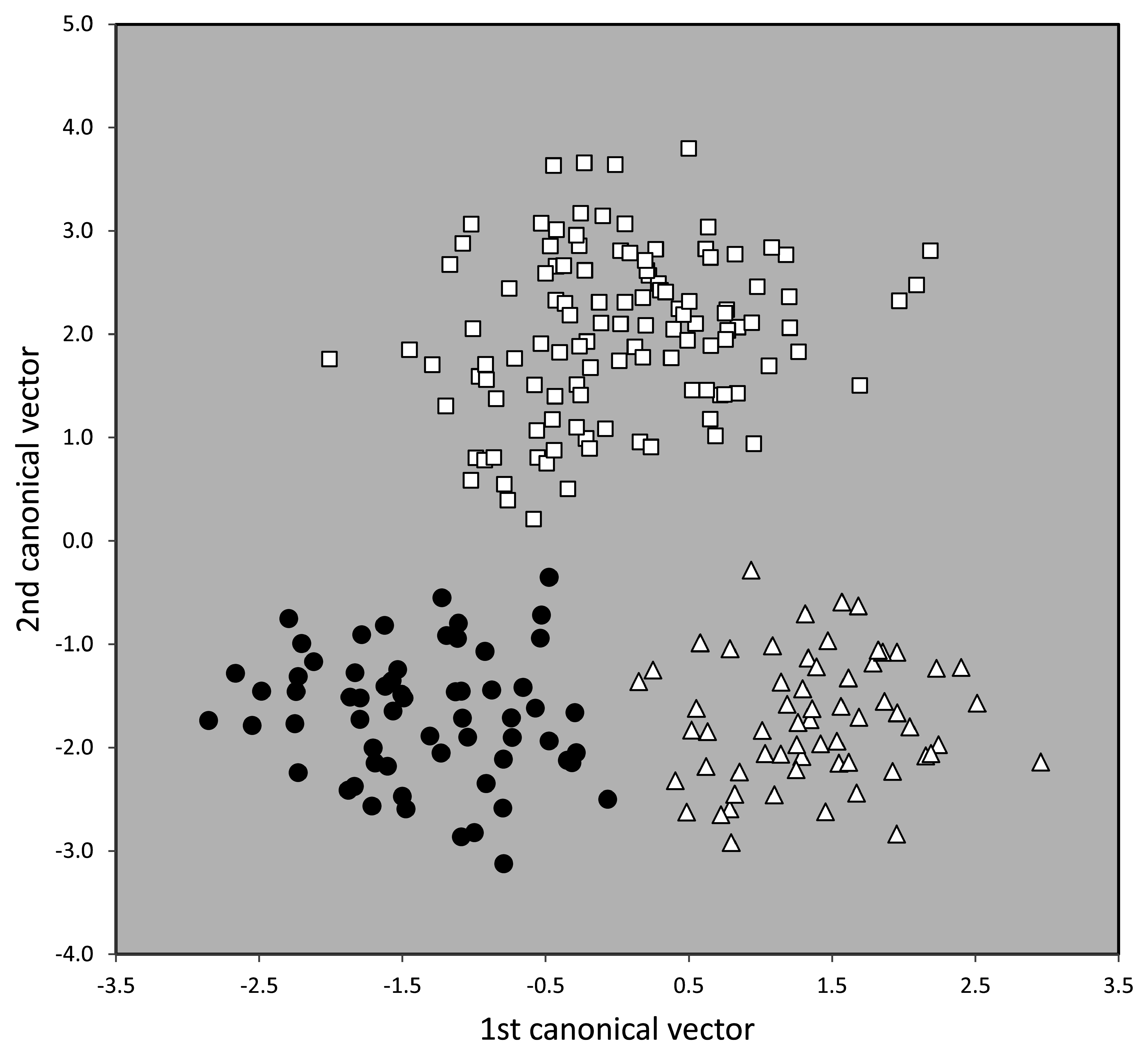
The second and third main cluster—altogether 240 nest samples and composed of the supercolonial species T. ibericum , T. dario i and T. magnum —were analyzed by NC-part.kmeans and NC-Ward. Both EDAs confirmed the existence of three clusters but disagreed in the classification of 8.8% of the samples. The species hypotheses of these unclear samples were then determined by wild-card runs in a LDA using all 15 characters. Relative to the controlling LDA, the misclassification by NC-part.kmeans was 4.9% in T. darioi , 3.2% in T. ibericum and 5.2% in T. magnum. The total disagreement of 4.4% over three entities justifies to hypothesize them as sufficiently divergent morphospecies. These hypotheses are strongly supported by running a 3-class LDA using 12 selected characters (nExCly 900, ML/CS 900 and MW/CS 900 were excluded) resulted in a mean classification error of 6.7% within 778 individuals but a complete separation on the nest sample level ( Fig.44 View FIGURE 44 ).
Block 2: Confirmation of NUMOBAT-based species hypotheses by genetic data
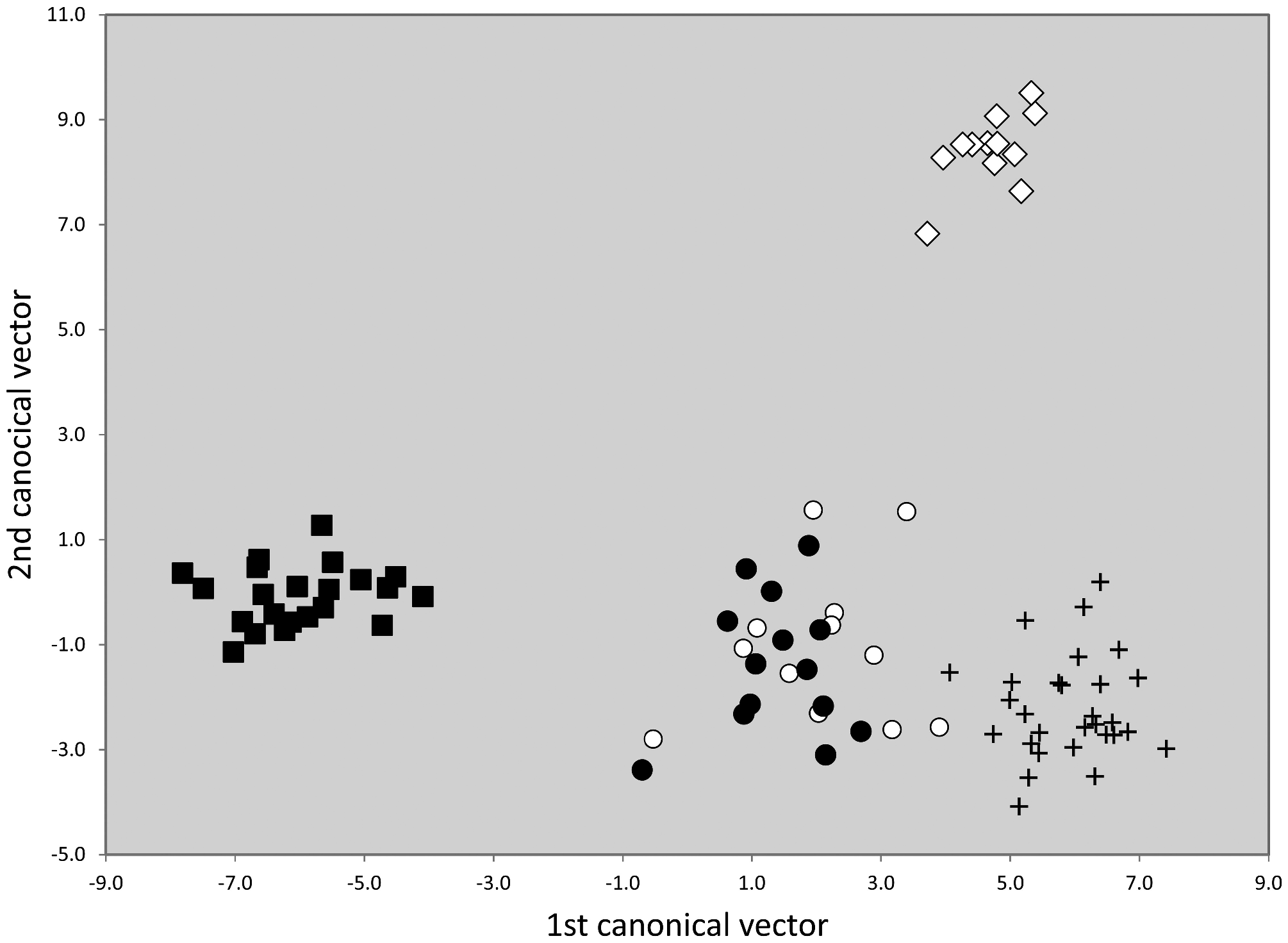
All five species clusters demonstrated by NUMOBAT could be confirmed by nuclear DNA. In a first step the first five principal components of microsatelite data of all 805 samples genotyped and belonging to this group were extracted. Then a LDA was run using these principal components as characters and and imposing the NUMOBAT classifications as hypotheses. Because the smallest class had only 12 samples and in order to avoid overfitting, the LDA used only the first four principal components. This procedure resulted in a very strong separation of T. nigerrimum and T. magnum and 100% agreement with NUMOBAT classification ( Fig.45 View FIGURE 45 ). On the other hand, this genetic data set completely failed to confirm the very strong NUMOBAT separation of T. hispanicum n. sp. and T. ibericum ( Fig. 42 View FIGURE 42 ) but provided a rather good separation of T. darioi from the collective cluster formed by T. hispanicum n. sp. and T. ibericum .
As next step of analysis the clearly separated T. magnum and T. nigerrimum samples were removed from the data set and the PCA of genetic data was run only with those 69 T. darioi , T. hispanicum and T. ibericum samples where both NUMOBAT and microsatellite data were available. A character-reduced LDA used only the PCA axes 1 and 3 as characters and removed axis 2 from the calculation as the latter only served to separate one individual having an unusual allele at one locus ( Fig. 46 View FIGURE 46 ). As result, 95.7% of NUMOBAT classifications in the 69 samples were confirmed by genetic data with one disagreement in each class. The posterior probabilities of the three controversial samples as calculated in wild-card runs of an LDA are shown in Tab. 4.
The Malaga sample is from a supercolony on the castle mountain near to the historic centre of Malaga Thus, colony structure and habitat speak against the genetic classification as T. hispanicum n. sp. The weak allocation to T. darioi by NUMOBAT data appears also rather unlikely since the natural range of T. darioi does not include S Spain. However, the anthropogenous dispersal capacity of this species should allow a transport to the south. Taking all indications together—zoogeography, colony structure and the combined posterior probabilities of NUMOBAT and genetic data—this sample should represent T. ibericum or a hybrid with participation of this species. The genetic classification of the sample from a supercolony in Rotterdam as T. hispanicum n. sp. is with a high probability wrong because the NUMOBAT signal pointing to T. ibericum is strong and because the former has never been introduced to areas outside Spain nor is it known to form supercolonies. The genetic classification of the sample from Lona de los Panaderos as T. darioi is also with certainty wrong because there is an extremely strong NUMOBAT indication as T. hispanicum n. sp. which is strongly supported by ecological indication: The altitude of this site ( 2300 m a.s.l.) is far above the upper altitudinal limit of any other species of the T. nigerrimum group—specifically T. darioi has never been found higher than 722 m.
Biology. Tapinoma nigerrimum frequently shows monodomous, not very large colonies which behave aggressively to each other. Polydomous colonies seem to exist but true supercoloniality or an invasive potential was not observed so far. Furthermore, in contrast to the supercolonial species, there is a trend in T. nigerrimum to dominate in semi-natural and rural habitats—without completely avoiding habitats with stronger anthropogenous impact ( Centanni et al. 2022). The rarity in coastal areas is probably a consequence of competition by the three supercolonial species. Alates were observed: 4 May ± 10 d [29 April–21 May] n = 5.
No known copyright restrictions apply. See Agosti, D., Egloff, W., 2009. Taxonomic information exchange and copyright: the Plazi approach. BMC Research Notes 2009, 2:53 for further explanation.
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |