Cothurnia salina, Zhuang, Yuan, Clamp, John C., Yi, Zhenzhen & Ji, Daode, 2016
|
publication ID |
https://doi.org/10.11646/zootaxa.4169.1.10 |
|
publication LSID |
lsid:zoobank.org:pub:0B9BA229-A1B1-4552-A933-9BFF363EE485 |
|
DOI |
https://doi.org/10.5281/zenodo.5619536 |
|
persistent identifier |
https://treatment.plazi.org/id/A34987A1-DE62-FFC7-FF71-3A5AE29785BB |
|
treatment provided by |
Plazi |
|
scientific name |
Cothurnia salina |
| status |
sp. nov. |
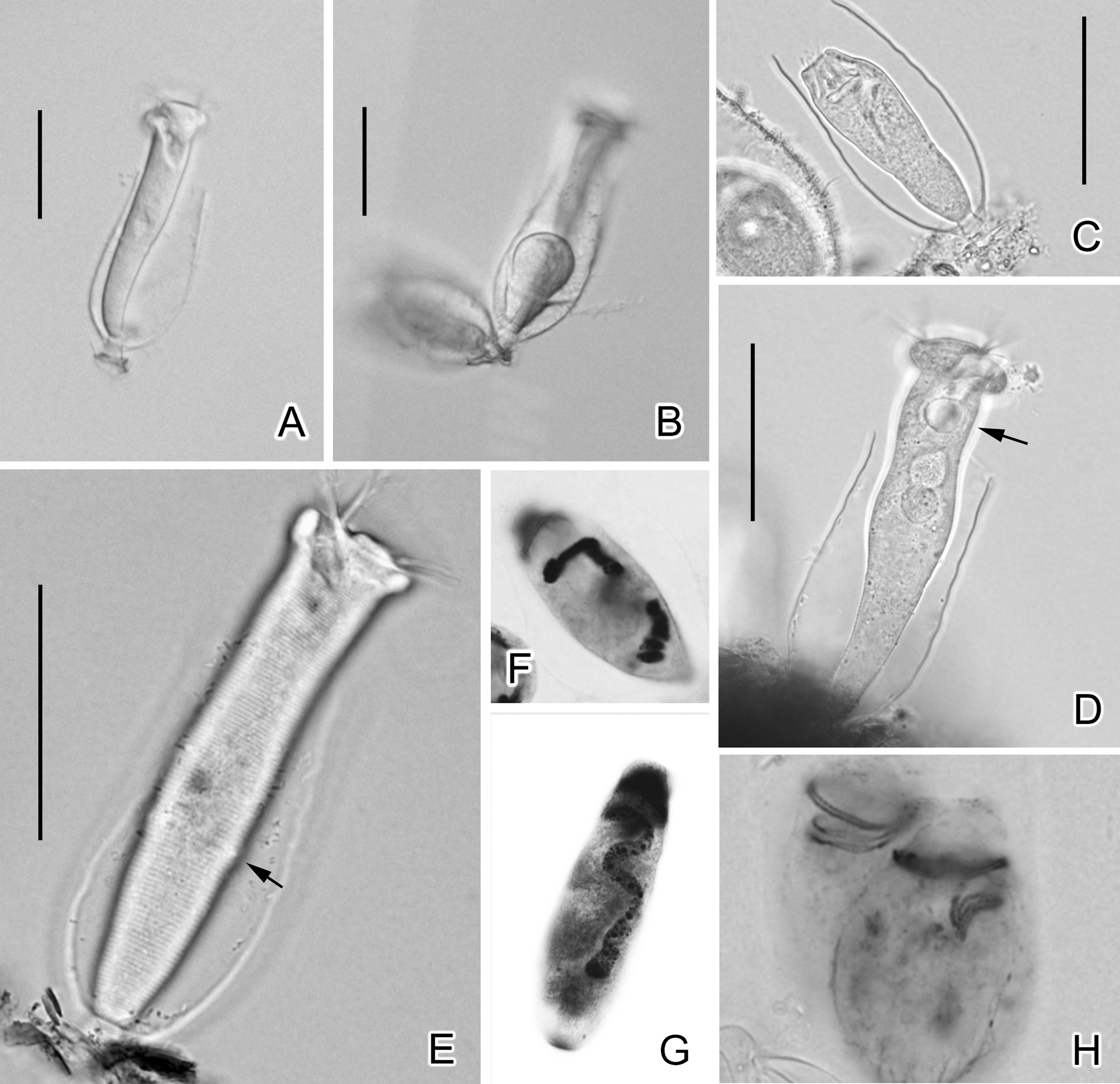
Morphology of Cothurnia salina n. sp.
( Figs. 1 View FIGURE 1 , 2 View FIGURE 2 ; Table 1 View TABLE 1 )
Diagnosis. Body elongated columnar, in vivo 80–98 × 12–19 µm; lorica barrel-shaped, with aboral part heavily thickened; stalk extremely short, with approximately ½ of its length within the lorica; macronucleus wormlike, longitudinally oriented; single contractile vacuole ventrally located; pellicle conspicuously striated, with 62–73 transverse and parallel striations between peristome and aboral trochal band, 32–38 between aboral trochal band and scopula; infundibular polykinety 3 (P3) consisting of two rows of kinetosomes, which are equal in length, parallel to each other and terminate adstomally between P1 and P2.
Type location. Muping Salt Factory , Yantai, China (37°25'33.80"N, 121°45'14.20"E). GoogleMaps
Type material. A slide (registration number 11102201 –01) containing the holotype specimen (protargol preparation, marked by ink circle) and a paratype slide with protargol-stained specimens (registration number 11102201 –02) have been deposited in the Laboratory of Protozoology, Ocean University of China .
Etymology. The specific epithet salina refers to the special hypersaline habitat of the new species.
Description. Solitary, with slender, cylindroid cell body measuring 80–98 × 12–19 µm ( Fig. 1 View FIGURE 1 A, 2A, D, E); body widest at the peristomial lip and not constricted below it ( Fig. 1 View FIGURE 1 A, 2A, E). Peristomial lip lacks medial infolding and measures 21–27 µm in diameter. Pellicular striae visible above ×400 magnification ( Fig. 2 View FIGURE 2 E), with 62–73 striations from peristome to aboral trochal band and 32–38 from aboral trochal band to scopula ( Table 1 View TABLE 1 ).
Cytoplasm colorless or slightly grayish, usually containing several food vacuoles (diameter 5–10 µm) located in center of body. Single contractile vacuole located adorally, beneath peristomial lip and near ventral wall of infundibulum ( Fig. 1 View FIGURE 1 A, 2D). Macronucleus slender, cylindroid, longitudinally oriented ( Fig. 1 View FIGURE 1 A, 2F, G); micronucleus not observed.
Lorica colorless, transparent, truncate pyriform, measuring 62–74 × 30–35 µm and aperture diameter of 21–30 µm. Aboral part of lorica thickened; wall 3 µm thick. Stalk 10 µm long, with approximately ½ of length exterior to lorica wall.
Oral infraciliature as shown in Fig. 1 View FIGURE 1 E, 2H. Haplokinety and polykinety make approximately one and one-half circuits together around peristome before entering infundibulum. Haplokinety and polykinety parallel to one another on peristome, diverging within infundibulum to lie on opposite walls ( Fig. 1 View FIGURE 1 E). Three infundibular polykineties (P1–3), with P1 and P2 consisting of three rows of kinetosomes and P3 of two rows. Adstomal ends of rows in P1 terminate at different levels, with inner row slightly shortened; P2 terminates adstomally above adstomal ends of P1 and P3, with row 3 not merging with P1 abstomally and significantly divergent from other two rows abstomally ( Fig. 1 View FIGURE 1 E). Rows of P3 parallel and equal in length, terminating adstomally at point approximately midway between adstomal ends of P2 and P1 ( Fig. 1 View FIGURE 1 E). Germinal kinety runs parallel to haplokinety in abstomal half of infundibulum ( Fig. 1 View FIGURE 1 E). Aboral trochal band consisting of series of dikinetids encircling cell at point 2/3 of distance from peristome to scopula ( Fig. 2 View FIGURE 2 E).
Phylogenetic analysis based on SSU rRNA sequences ( Fig. 3 View FIGURE 3 ). The SSU rRNA gene sequence of Cothurnia salina (GenBank accession number KT956998 View Materials ) is 1,629 bp long and has a GC content of 43.09%. The sequence of C. salina differs from that of C. annulata ( KU363275 View Materials ) by 38 nucleotides (sequence identity 97.4%); C. sp. 0 924 ( KU363268 View Materials ) by 69 nucleotides (sequence identity 95.2%).
Topologies of BI and ML trees were basically congruent, and thus, only the topology of the BI tree is presented in Fig. 3 View FIGURE 3 , with support values from both analyses indicated on branches. In both BI and ML trees, C. salina n. sp. clustered in the family Vaginicolidae as expected. C. salina n. sp. clustered with C. annulata with full support (1.00 BI, 100% ML) and these two species are sisters to the species C. sp. 0 924 with moderate support (0.80 BI, 57% ML).
All data based on specimens in vivo.
Abbreviations: Min, minimum; Max, maximum; Mean, arithmetic mean; SD, standard deviation; CV, coefficient of variation (%); n, number of individuals investigated.
TABLE 1. Morphometric characteristics on Cothurnia salina n. sp.
| Characters | Min | Max | Mean | SD | CV | n |
|---|---|---|---|---|---|---|
| Body length (µm) | 80 | 98 | 92.6 | 5.4 | 29.0 | 12 |
| Body width (µm) | 12 | 19 | 16.3 | 2.5 | 6.2 | 15 |
| Diameter of peristomial lip (µm) | 21 | 27 | 23.3 | 1.8 | 3.3 | 14 |
| Lorica length (µm) | 62 | 74 | 67.8 | 4.1 | 16.8 | 10 |
| Lorica width (µm) | 30 | 35 | 32.2 | 1.4 | 2.0 | 11 |
| Diameter of lorica aperture (µm) | 21 | 30 | 25.7 | 2.9 | 8.2 | 11 |
| Number of striations, peristome to aboral trochal band | 62 | 73 | 67 | 4.5 | 20.6 | 7 |
| Number of striations, aboral trochal band to scopula | 32 | 38 | 35 | 2.0 | 3.9 | 7 |
No known copyright restrictions apply. See Agosti, D., Egloff, W., 2009. Taxonomic information exchange and copyright: the Plazi approach. BMC Research Notes 2009, 2:53 for further explanation.
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |