Aplectana xishuangbannaensis, Chen & Gu & Ni & Li, 2021
|
publication ID |
https://doi.org/ 10.1186/s13071-021-04667-9 |
|
DOI |
https://doi.org/10.5281/zenodo.11109894 |
|
persistent identifier |
https://treatment.plazi.org/id/BA5287B6-FFD8-FFA8-FF2F-FC0B8E718254 |
|
treatment provided by |
Felipe |
|
scientific name |
Aplectana xishuangbannaensis |
| status |
sp. nov. |
Aplectana xishuangbannaensis View in CoL n. sp.
Type host: White-spotted thigh tree-frog Polypedates megacephalus (Hallowell) ( Anura : Rhacophoridae ).
Type-locality: XiShuangBanNa Tropical Botanical Garden (21°41 ′ N, 101°25 ′ E), Yunnan Province, China GoogleMaps .
Type specimens: Holotype: male ( HBNU–N-2020A009L ); allotype: female ( HBNU–N-2020A010L ); paratypes: 41 males, 122 females ( HBNU–N-2020A011L ).
Site of infection: Intestine.
Prevalence and intensity of infection: 12.1% (11 P. megacephalus infected out of 91 examined) were infected with intensity of 1–88 (mean 15.0) nematodes.
ZooBank registration: To comply with the regulations set out in Article 8.5 of the amended 2012 version of the International Code of Zoological Nomenclature ( ICZN) [ 19], details of the new species have been submitted to ZooBank. Te Life Science Identifier ( LSID) of the article is urn:lsid:zoobank.org:pub:09F4B1EF-C3AF42E6-80E6-B734D6B084B8. Te LSID for the new name Aplectana xishuangbannaensis is urn:lsid:zoobank.org:act:5E4C6C18-7B72-4C28-BD28-6964C6D8F0A3 .
Etymology: Te specific epithet refers to the type location XiShuangBanNa Tropical Botanical Garden, Yunnan Province, China.
Description
General
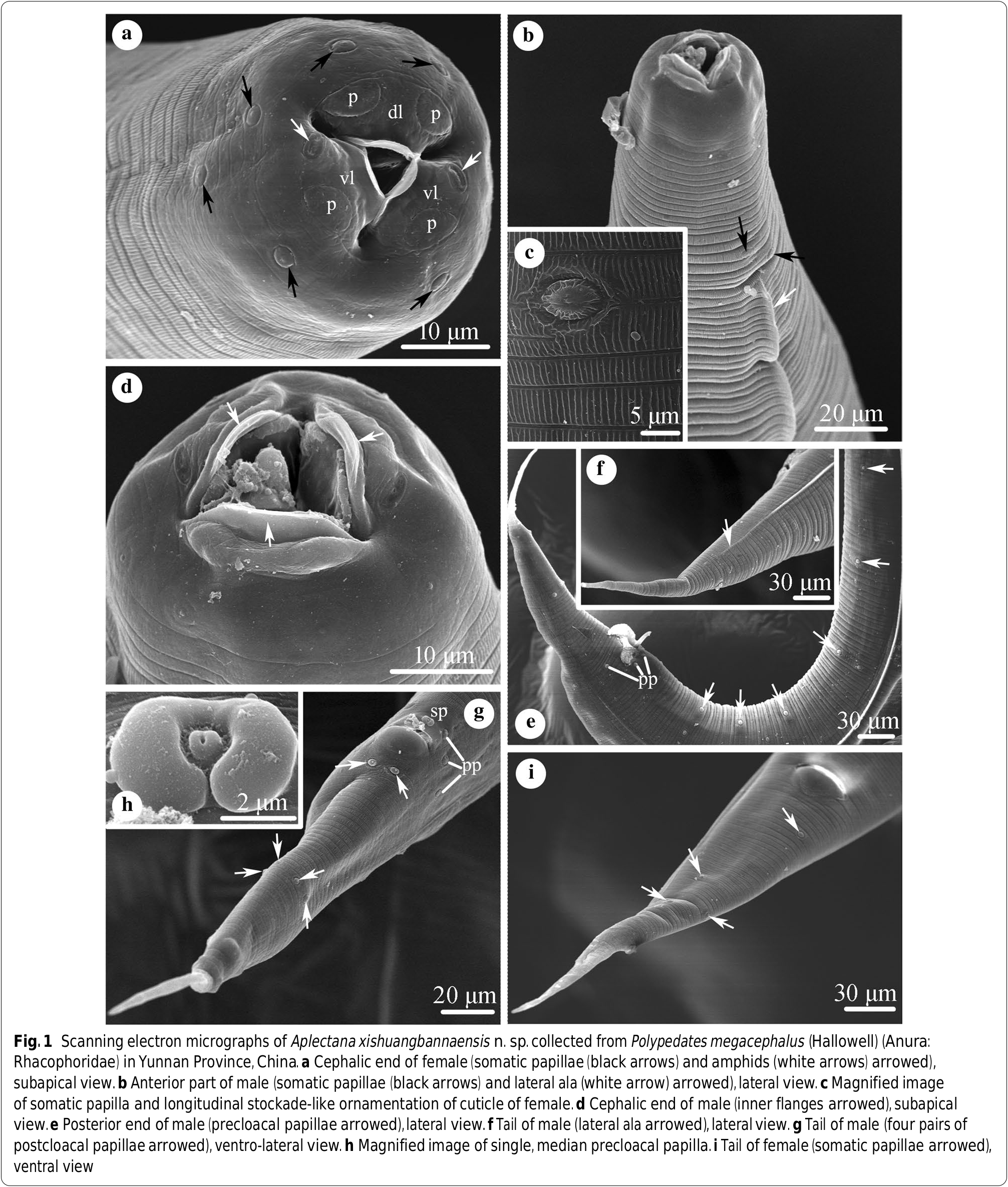
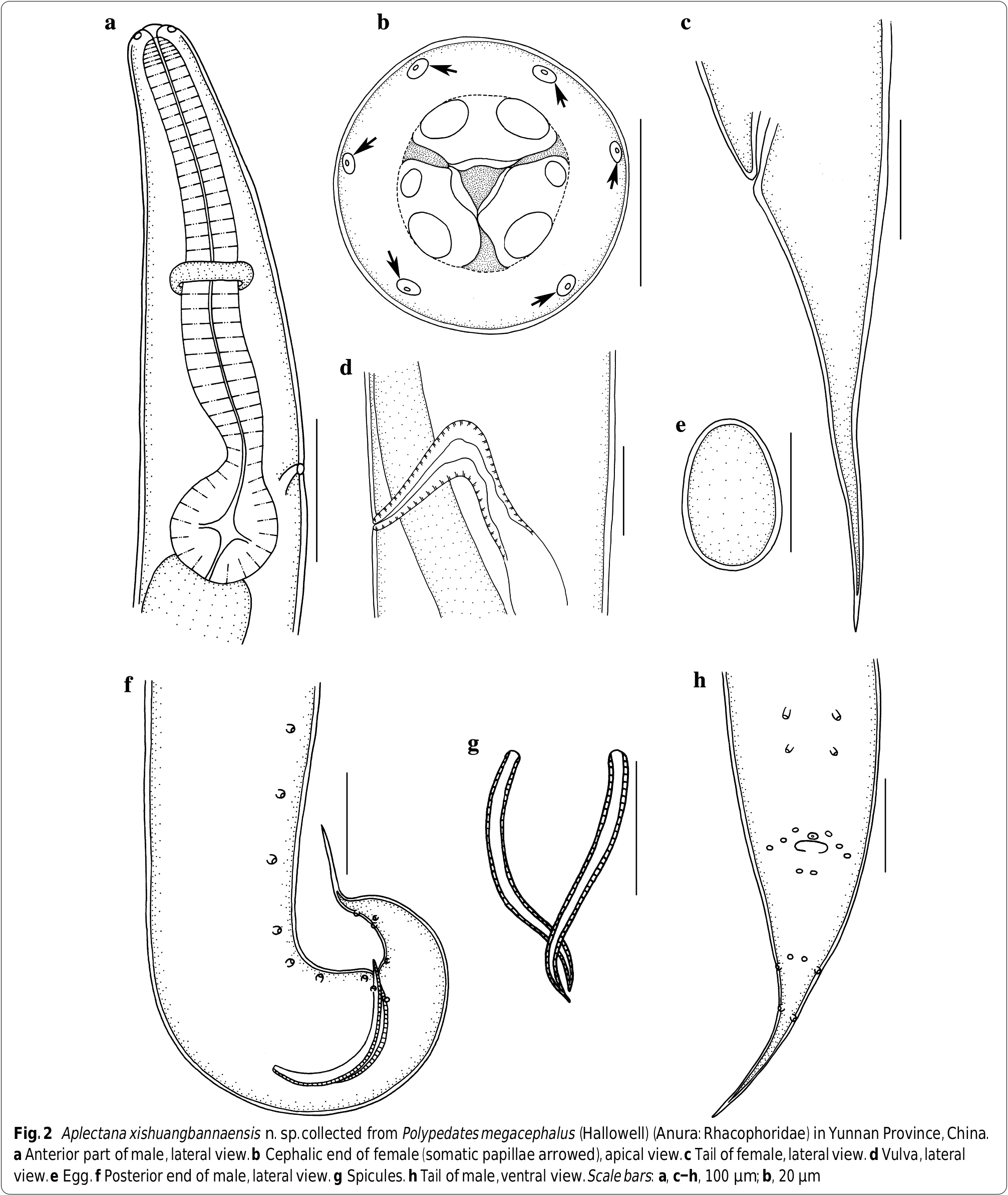
Small-sized, whitish nematodes. Body cylindrical, maximum width at about region of middle body. Cuticle with fine transverse striations and longitudinal stockade-like ornamentation ( Fig. 1a–c View Fig ). Somatic papillae small, distributed irregularly over body surface ( Figs. 1a–c, e, i View Fig , 2b View Fig ). Lateral alae extending from 60–70 posterior to base of lips as far as about middle of tail in both sexes ( Fig. 1b, f, i View Fig ). Oral aperture simple, triangular, surrounded by 3 small lips, each with inner flanges ( Figs. 1a, b, d View Fig , 2b View Fig ). Dorsal lip with pair of large double cephalic papillae; subventral lips with single large double cephalic papilla and amphid each ( Figs. 1a View Fig , 2b View Fig ). Oesophagus divided into anterior short pharynx, cylindrical corpus, slightly narrower isthmus and terminal posterior bulb with valves ( Fig. 2a View Fig ). Nerve ring located at about 1/2 of oesophageal length. Excretory pore slightly anterior to of oesophageal bulb ( Fig. 2a View Fig ). Tail of both sexes conical, with long filamentous tip ( Figs. 1e–g, i View Fig , 2c, f, h View Fig ).
Male
[Based on 10 mature specimens; Figs. 1b, d–h View Fig , 2a, f–h View Fig ]: Body 2.32–2.72 (2.49) mm long, maximum width 139–178 (158). Oesophagus 317–426 (374) long, representing 12.6–16.1 (15.0) % of body length; pharynx + corpus + isthmus 248–356 (307) long, bulb 59–69 (67) × 50–59 (54) ( Fig. 2a View Fig ). Nerve ring 158–198 (176) and excretory pore 257–376 (334) from anterior extremity, respectively ( Fig. 2a View Fig ). Posterior end of body distinctly curved ventrally ( Figs. 1e View Fig , 2f View Fig ). Spicules small, similar in shape and length, 139–178 (161) long, distal end pointed, representing 5.98–7.09 (6.47) % of body length ( Fig. 2g View Fig ). Gubernaculum absent. Caudal papillae: 6 pairs of precloacal, 3 pairs paracloacal (distinguishable from somatic papillae) and 4 pairs postcloacal papillae. Single median, ventral precloacal papilla present ( Figs. 1g, h View Fig , 2h View Fig ). Tail 198–248 (230) long, representing 8.26 ‒ 9.84 (9.26) % of body length ( Figs. 1e–g View Fig , 2f, h View Fig ).
Female
[Based on 10 mature specimens; Figs. 1a, c, i View Fig , 2b–e View Fig ]: Body 3.54–3.86 (3.65) mm long, maximum width 248–297 (272). Oesophagus 416–446 (431) long, representing 11.0–12.6 (11.8) % of body length; pharynx + corpus + isthmus 347–366 (356) long, bulb 69–79 (74) × 50–69 (62). Nerve ring 208–228 (215) and excretory pore 347–386 (366) from anterior extremity, respectively. Vulva transverse slit, 1.60–2.10 (1.89) mm from anterior extremity, at 44.8–54.5 (51.8) % of body length. Ovaries two, located anterior to vulva. Vagina muscular ( Fig. 2d View Fig ). Uteri amphidelphic, full of eggs in different stages of development; egg oval, large, with smooth surface, 149–297 (205) × 99–238 (146) (n = 20) ( Fig. 2e View Fig ). Tail 347–406 (384) long, representing 9.78 ‒ 11.1 (10.5) % of body length ( Figs. 1i View Fig , 2c View Fig ).
Genetic characterization
Partial 18S region
Tree 18S sequences of Aplectana xishuangbannaensis n. sp. (accession numbers MW329041–MW329043) obtained were all 1539 bp long, representing only one genotype. Tere is no species of Aplectana with 18S sequenced registered in GenBank. Pairwise comparison between A. xishuangbannaensis n. sp. and the other species of Cosmocercidae regarding the 18S sequences available in GenBank, including Cosmocerca simile (MN839758–MN839760), Cosmocercoides dukae (FJ516753), C. pulcher (LC018444, MH178322– MH178326), C. qingtianensis (MH032769–MH032771, MH178319–MH178321), C. tonkinensis (AB908160), C. wuyiensis (MK110872), Nemhelix bakeri (DQ118537) and Raillietnema sp. (DQ503461), displayed 1.88– 3.77% nucleotide divergence.
Partial ITS-1 region
Tree ITS-1 sequences of A. xishuangbannaensis n. sp. (accession numbers MW329035–MW329037) obtained were all 554 bp long, representing only one genotype. Tere are two species of Aplectana with ITS sequences available in GenBank, including A. chamaeleonis (MN907375 ‒ MN907378) and Aplectana sp. ’ Neyraplectana ’ PNLS-530 (MH836325). Pairwise comparison between A. xishuangbannaensis n. sp. and the previously mentioned taxa showed 46.67 and 45.47% nucleotide divergence, respectively. Pairwise comparison between A. xishuangbannaensis n. sp. and the other species of Cosmocercidae regarding the ITS sequences available in GenBank, including Cosmocerca japonica (LC052772 ‒ LC052782), C. longicauda (MG594349 ‒ MG594351), C. ornata (MT108302), Cosmocerca sp. LL-2020 (MT108303), C. simile (MN839761 ‒ MN839768), Cosmocercoides pulcher (MH178314–MH178318, LC018444), C. qingtianensis (MH178311–MH178313, MH032772–MH032774), C. tonkinensis (AB908160, AB908161) and C. wuyiensis (MK110871), displayed 28.53–47.52% of nucleotide divergence.
Partial 28S region
Tree 28S sequences of A. xishuangbannaensis n. sp. (accession numbers MW329038–MW329040) obtained were all 740 bp long, representing only one genotype. Tere is only one species of Aplectana , Aplectana sp. ’ Neyraplectana ’ PNLS-530, with 28S sequence data (MH909070) available in GenBank. Pairwise comparison between A. xishuangbannaensis n. sp. and the previously mentioned taxon showed 20.67% of nucleotide divergence. Pairwise comparison between A. xishuangbannaensis n. sp. and the other species of Cosmocercidae with 28S sequences available in GenBank, including Cosmocerca simile (MN839755–MN839757), Cosmocercoides pulcher (LC018444) and C. tonkinensis (AB908160), displayed 16.78–17.94% of nucleotide divergence.
Partial cox1 region
Tree cox 1 sequences of A. xishuangbannaensis n. sp. (accession numbers MW327586–MW327588) obtained were all 384 bp long, representing only one genotype. Tere is no species of Aplectana with cox 1 sequence registered in GenBank. Pairwise comparison between A. xishuangbannaensis n. sp. and the other species of Cosmocercidae regarding the cox 1 sequences available in GenBank, including C. japonica (LC052756 ‒ LC052770), C. ornata (MT108304), Cosmocerca sp. LL-2020 (MT108305), C. simile (MN833301 ‒ MN833303), C. pulcher (MH178306–MH178310, LC052771) and C. qingtianensis (MH178303–MH178305, MH032775– MH032777), displayed 10.23–21.09% nucleotide divergence.
Phylogenetic analyses
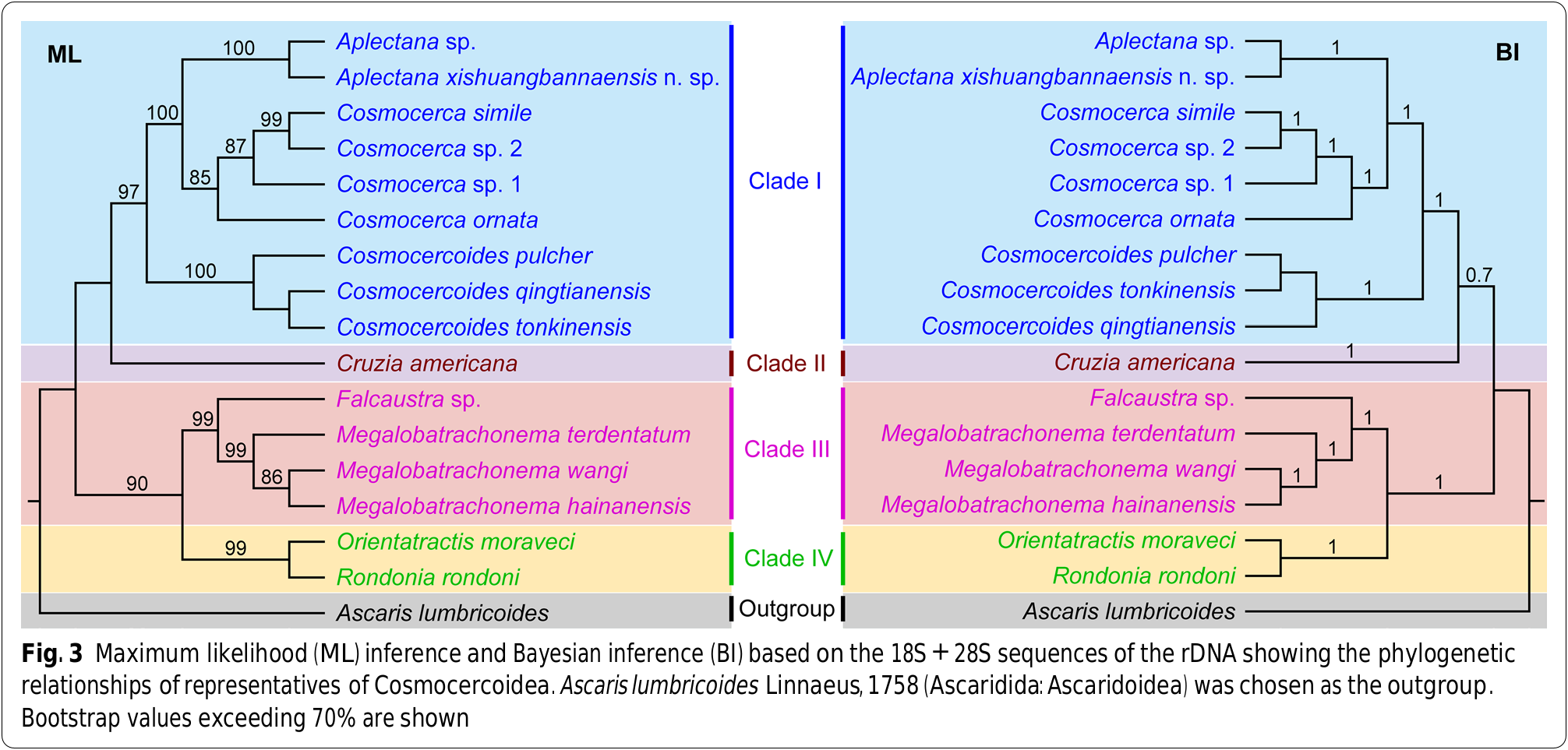
Phylogenetic trees inferred from maximum likelihood (ML) and Bayesian inference (BI) showed that representatives of Cosmocercoidea were divided into four major clades ( Fig. 3 View Fig ). Clade I included the species of three genera Cosmocerca , Cosmocercoides and Aplectana , representing the family Cosmocercidae . Among the three genera, Cosmocerca displayed a closer relationship to Aplectana rather than Cosmocercoides . Clade II included only Cruzia americana (a common nematode parasite in the digestive tract of opossums), which belongs to the subfamily Cruzinae in the family Kathlaniidae according to the current classification [ 1]. Clade III included species of Falcaustra and Megalobatrachonema , which represent the family Kathlaniidae . Te representatives of Orientatractis and Rondonia formed Clade IV, representing the family Atractidae .
No known copyright restrictions apply. See Agosti, D., Egloff, W., 2009. Taxonomic information exchange and copyright: the Plazi approach. BMC Research Notes 2009, 2:53 for further explanation.
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |