CLADOPATHIDAE KINOSHITA, 1910
|
publication ID |
https://doi.org/10.1111/zoj.12060 |
|
persistent identifier |
https://treatment.plazi.org/id/DE793A5A-FFB7-ED4E-1392-FB8082B7FC41 |
|
treatment provided by |
Marcus |
|
scientific name |
CLADOPATHIDAE KINOSHITA, 1910 |
| status |
|
CLADOPATHIDAE KINOSHITA, 1910 View in CoL
The family Cladopathidae (six genera, 18 species) is characterized by the presence of six mesenteries in the polyps, and is the only antipatharian family in which the polyps have only primary mesenteries and no secondary mesenteries. Colonies are always pinnulated and may be monopodial or branched to varying degrees. Corallum spines are simple, smoothsurfaced, and conical to acicular (i.e. needle-like). The polyps are greatly elongated transversely along the skeletal axis, up to about 6 mm (but up to 9 mm in the Hexapathinae), and are similar in size and shape to polyps in the Schizopathidae . The family is divided into three subfamilies – the Cladopathinae (three genera), Hexapathinae (two) and Sibopathinae (one) – distinguished based on the degree of branching of the corallum, number of rows of primary pinnules, and presence/absence of an actinopharynx ( Opresko, 2003b; Opresko & de Laia Loiola, 2008).
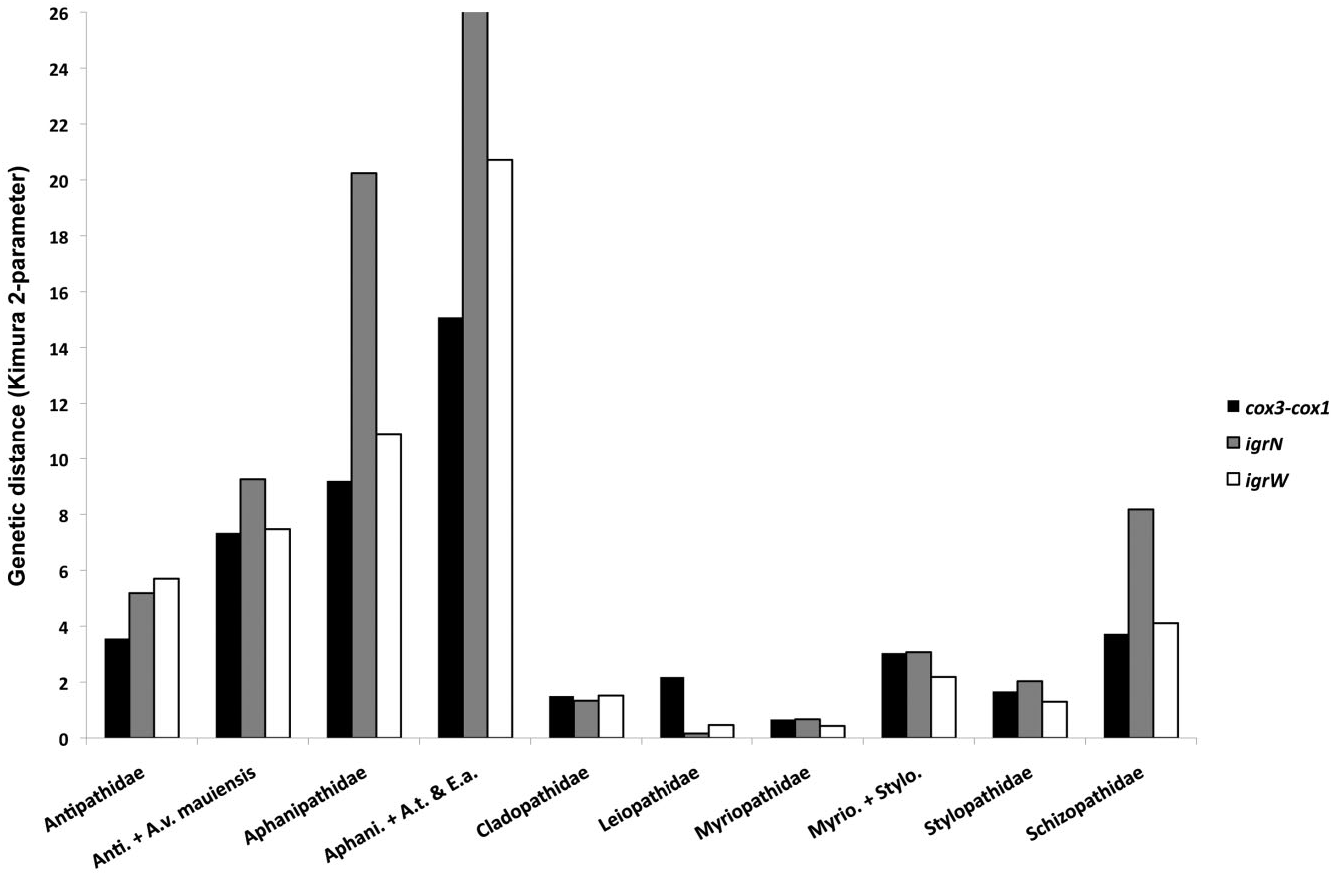
Based on morphological analyses, six species, representing four genera (and all three subfamilies), were obtained for sequencing, of which four could be referred with certainty to valid nominal species ( Table 1 & Supporting information Table S1). In the mt-contig phylogenies (only a single species was included in the nuc-contig alignment), all species grouped in a single clade with the exception of the cladopathid Sibopathes macrospina , which grouped within the Schizopathidae , thereby rendering Cladopathidae polyphyletic. The remaining species were divided into two subclades separated by a genetic distance (K2P) of < 1.5% ( Fig. 1 View Figure 1 ); the subclades did not reflect subfamily designations. Indeed, among the colonies examined for this family, several could not be distinguished using mtDNA even though they belonged to separate genera or species.
One of the two subclades comprised Chrysopathes species and three haplotypes were differentiated [as only igrW was obtained for an undescribed species of Chrysopathes (2330-AB7), this specimen was not included in the mt-contig phylogeny; however, based on the igrW phylogeny, it grouped with C. formosa (41-99-6)]. Among the seven C. formosa colonies sequenced, two mt-contig haplotypes were observed (represented in the phylogenies by 41-99-6 from the Gulf of Alaska and DQ304771 View Materials from Fieberling Guyot), differentiated by only a single substitution across 1025 sites compared (at cox 3-igrC - cox 1). Two mt-contig haplotypes were also observed among the five C. speciosa colonies (all from the Gulf of Alaska), but four of these could not be differentiated from the Alaskan C. formosa haplotypes; only C. speciosa specimen 41-1A-6 (also from Alaska) differed from Alaskan C. formosa (at igrW, but not igrN; based on a single substitution across 630 bp). Given the extremely low genetic divergence among the Chrysopathes haplotypes, we suggest that either C. formosa and C. speciosa are very closely related species, or a single species with intraspecific variation. The morphological distinction of C. formosa from C. speciosa is based on limited differences: primarily the number and density of subpinnules ( Opresko, 2003b). Three additional species have been described for the genus Chrysopathes , including two from the Atlantic ( Opresko & de Laia Loiola, 2008), and sequences from these will help evaluate the relationship of mtDNA variation and species status in this genus.
The second subclade comprised the genera Trissopathes Opresko, 2003b and Heteropathes Opresko, 2011 (previously Heliopathes Opresko, 2003b ). As with Chrysopathes , haplotypes were shared across nominal taxa, but in this case it is across genera. We sequenced two colonies of Trissopathes pseudotristicha Opresko, 2003b – the holotype (USNM 98848) from Hawaii ( 432 m depth) and a specimen (J 2095-2-1-3) from the Aleutian Islands, Alaska ( 2827 m); both differed at all three mt gene regions sequenced. However, the mt-contig haplotype of the Alaskan specimen was identical to that of Heteropathes sp. (T886-A8) collected from off the south-western coast of Oregon ( 3030 m). All three specimens were identified by D.M.O., who also described these two genera ( Opresko, 2003b), so we are confident that they fit the generic diagnoses. Additionally, DNA was re-extracted, PCR amplified and sequenced for both Heteropathes sp. (T886-A8) and T. pseudotristicha (J 2095-2-1-3) to negate the possibility of a mix-up in the lab.; results were confirmed. The lack of genetic differentiation suggests either slow genetic divergence at these gene regions, hybridization with subsequent introgression, or a closer relationship than is suggested by their separate generic distinctions. There are three described species of Heteropathes , distributed in the Gulf of Mexico, Indian Ocean, and North Pacific, and three of Trissopathes , distributed in the North and South Pacific and Southern Ocean. Additional sequences from at least some of these species will be required to fully understand the status of these genera. With the exception of S. macrospina , the remaining cladopathids grouped sister to the Schizopathidae with strong support (BS: 99.7–99.9; BPP: 100).
Two individuals of Sibopathes macrospina from the Gulf of Mexico shared the same haplotype with five individuals from North Atlantic seamounts (Supporting information Table S1; the latter specimens were not analysed histologically to confirm the presence of only six mesenteries). Based on the mt-contig phylogenies, S. macrospina groups within a larger clade of Parantipathes spp. (family Schizopathidae ), the latter of which were also collected from North Atlantic seamounts ( Fig. 2 View Figure 2 ). This finding was not completely unexpected, as Sibopathes van Pesch, 1914 shares similar skeletal characteristics with Parantipathes , as well as having transversely elongated polyps of relatively small size (generally < 3 mm) ( Opresko, 1993). Members of the family Cladopathidae are defined by polyps that have six primary mesenteries and no secondary mesenteries. Sibopathes was afforded its own subfamily within the Cladopathidae due to the presence of what van Pesch (1914) described as numerous primitive characters, such as the lack of an actinopharynx, and thus six incomplete primary mesenteries, and absence of mesenteric filaments ( Opresko, 1993). The placement of S. macrospina within a larger clade of Parantipathes spp. suggests that these morphological characters were secondarily lost, possibly as a result of the extreme transverse elongation of the polyp ( Opresko, 1993); thus, these ‘primitive’ characteristics are actually derived. Based on preliminary observations by M.R.B., and referring specifically to colonies from north-west Atlantic seamounts, Sibopathes can be readily distinguished from Parantipathes based on branching pattern alone; Sibopathes is highly branched while Parantipathes is either monopodial or contains a single forking branch. The genus Sibopathes contains two species, S. macrospina and S. gephura van Pesch, 1914 , the latter of which is the type species. These two species differ primarily in the orientation and arrangement of the pinnules, the size of the spines, and the diameter of the central axial canal in the pinnules ( Table 4). Thus, molecular data must also be obtained for S. gephura before reassessing the current classification of the subfamily Sibopathinae , and its sole genus Sibopathes .
Although containing only a single cladopathid ( T. pseudotristicha USNM 98848), the results of the nuc-contig phylogenies were congruent with the results of the mt-contig phylogenies, with the single cladopathid grouping sister to the Schizopathidae with strong support (BS: 96.1; BPP: 99–100).
No known copyright restrictions apply. See Agosti, D., Egloff, W., 2009. Taxonomic information exchange and copyright: the Plazi approach. BMC Research Notes 2009, 2:53 for further explanation.