Isleria, Bravo, Gustavo A., Chesser, Terry & Brumfield, Robb T., 2012
|
publication ID |
https://doi.org/10.5281/zenodo.208867 |
|
DOI |
https://doi.org/10.5281/zenodo.5658711 |
|
persistent identifier |
https://treatment.plazi.org/id/E1458793-2145-FF82-FF29-F969FD20FC1C |
|
treatment provided by |
Plazi |
|
scientific name |
Isleria |
| status |
gen. nov. |
Isleria View in CoL gen. nov.
Type species. Myrmothera guttata Vieillot , ca. 1825.
Included species. Isleria guttata (Vieillot, ca. 1825) comb. nov., Rufous-bellied Antwren; Isleria hauxwelli ( Sclater, 1857) comb. nov., Plain-throated Antwren.
Diagnosis, morphology. Small birds of the family Thamnophilidae typically 8.5–11g and 8.5–9.5 cm long ( Zimmer & Isler, 2003); rectrix 1 length 22–24.5 mm; rectrix 1 width 5.3–6.3 mm; wing chord 50–53 mm; primary 10 length 33–37 mm; secondary 1 length 43–46.5 mm; bill length from nares to tip 8.5–9.5 mm; bill width at nares 3–4 mm; bill depth at nares 3.3–3.8 mm; tarsus length 18–20 mm; hallux length 11.5–13 mm (measurements from LSUMZ 84823, 109924, 109926, 161753, 165714; MPEG 45899, 45900, 51034). Sexually dimorphic in plumage, males predominantly gray and females brown. Distinguished from all other small antwrens primarily by the presence of conspicuous white or buffy distal markings on the tertiary feathers of both males and females. The lack of any plumage markings on the head, underparts, and upperparts of either sex also makes Isleria fully diagnosable from Myrmotherula gularis Spix , the stipple-throated antwren assemblage ( Epinecrophylla ), and the streaked antwren assemblage ( Myrmotherula, sensu Isler et al. 2006 ). Isleria species have longer and larger tarsi, feet, and toes than other species of Myrmotherula except M. gularis , and shorter tails than all other antwrens except those in the streaked assemblage. The lack of markings on the breast and back makes Isleria diagnosable from other small antbirds with conspicuous distal spots on the tertiaries, such as Hylophylax Ridgway and Dichrozona Ridgway , and its smaller size distinguishes Isleria from Megastictus Ridgway ( 18–21 g; Zimmer & Isler, 2003).
Etymology. We take great pleasure in naming this genus after Morton and Phyllis Isler in recognition of their outstanding contributions to the knowledge of the taxonomy and systematics of Neotropical birds, especially antbirds of the family Thamnophilidae . Their milestone syntheses of tanager biology ( Isler & Isler, 1987), antbird vocalizations ( Isler & Whitney, 2002), and antbird biology ( Zimmer & Isler, 2003) have provided solid foundations for the advancement of scientific knowledge of these groups. In addition, their numerous studies of vocal variation in antbirds (e.g. Isler et al., 1997, 1999, 2001, 2005, 2007; Isler & Whitney, 2011) have not only improved our understanding of species limits within the group, but have provided methodological guidelines to incorporate vocal and geographic data to better understand taxonomy, systematics, and evolutionary processes in the family ( Isler, 1997; Isler et al., 1998). The Islers’ enthusiasm to learn, collaborate, and share their extraordinary knowledge of bird biology has become their trademark, and made them role models for upcoming generations of ornithologists. The name Isleria is feminine in gender.
Molecular analyses. Here, we present a subset of taxa from a densely sampled molecular phylogeny of the Thamnophilidae (including 214 of 220 species) to show that I. guttata and I. hauxwelli are only distantly related to the Myrmotherula clade. Taxon sampling ( Table 1 View TABLE 1 ) for this subset includes 39 individuals representing 28 species and 19 genera (13% and 40% of the family, respectively), and includes samples of the following species included in or recently split from Myrmotherula : M. brachyura (2; type species), M. multostriata Sclater (1), M. axillaris Vieillot (2), M. menetriesii Orbigny (1), M. gularis (2), Epinecrophylla haematonota Sclater (1; type species), E. erythrura Sclater (1), Isleria guttata (5), and I. hauxwelli (5). Samples of Isleria cover the full extent of the distribution of both species and represent all subspecies currently recognized. For outgroups we used Chamaeza campanisona Lichtenstein ( Formicariidae ; UWBM KGB14), Hylopezus berlepschi Hellmayr ( Grallariidae ; FMNH 322345), and Melanopareia elegans Lesson ( Melanopareiidae ; LSUMZ B-5245/5246), and the tree was rooted between ingroup and outgroup.
Total DNA was extracted from 25 mg of pectoral muscle using the Qiagen DNeasy kit, following the manufacturer’s protocol. Based on the methods described in Brumfield et al. (2007) we amplified and sequenced three mitochondrial genes (NADH dehydrogenase subunit 2 – ND2, 1041 bp; NADH dehydrogenase subunit 3 – ND3, 351 bp; cytochrome b – cytb, 842 bp) and one autosomal nuclear intron (β -fibrinogen intron 5 – βF5, 554 bp). We also amplified two coding nuclear genes ( recombination activation gene 1 – RAG1, 2872 bp; recombination activation gene 2 – RAG2, 1142 bp) following the methods described in Groth and Barrowclough (1999) and Barker et al. 2002. Additionally, some sequences were obtained from previous publications of our own work ( Brumfield & Edwards, 2007; Brumfield et al., 2007; Moyle et al., 2009; Gómez et al., 2010). Analyses were conducted using a concatenated six-gene alignment containing 6802 bp.
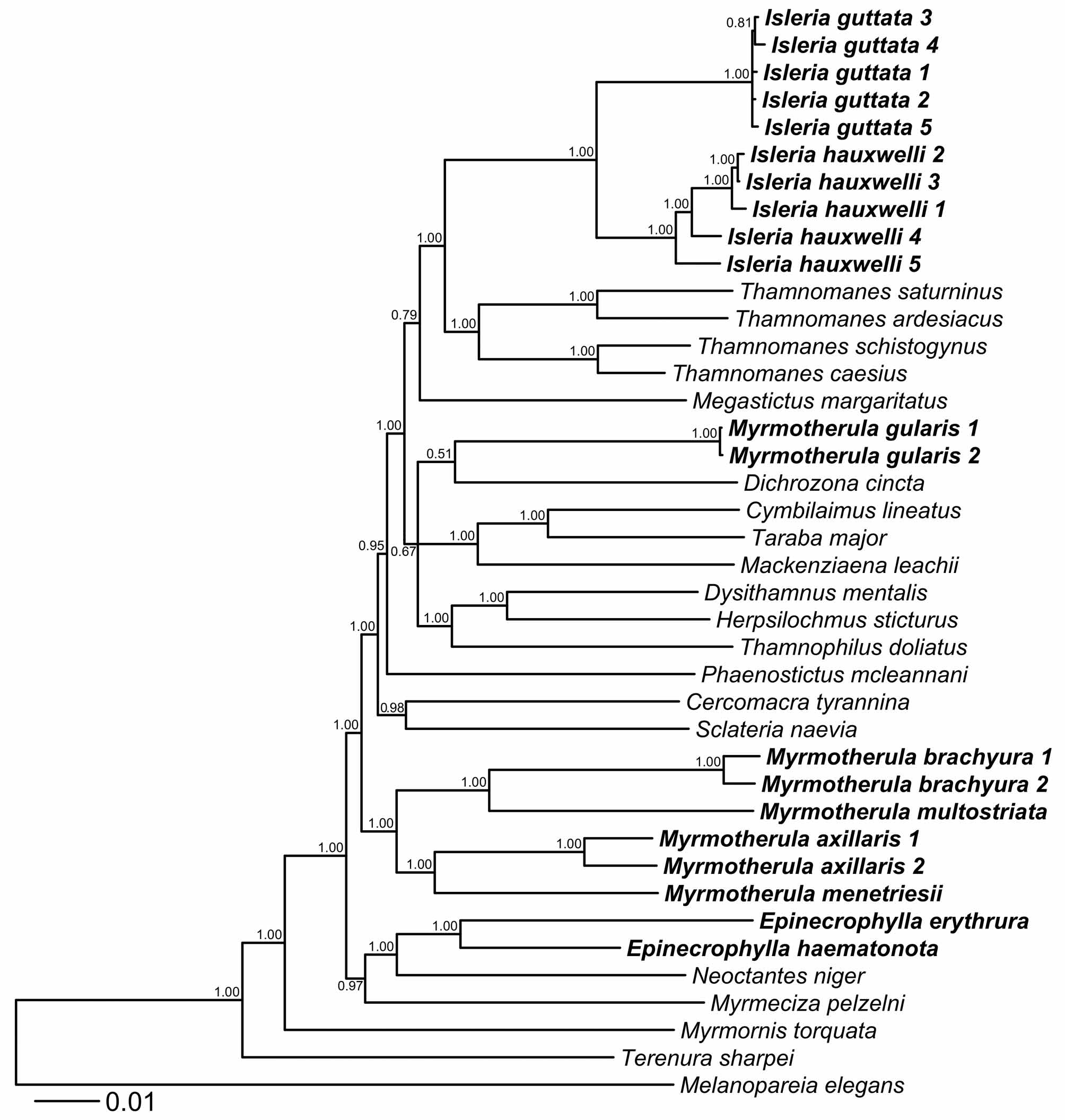
To avoid the risk of over-parameterizing our estimate of phylogeny, we ran ML analyses for six different partition schemes under the GTR+Γ model of nucleotide substitution using RAxML 7.2.7 ( Stamatakis, 2006) on the Cipres Science Gateway V 3.1 ( Miller et al., 2010). We then calculated the Akaike Information Criterion (AIC) for each partition and established that the most informative scheme is the fully partitioned dataset (16 partitions; the nuclear intron and each codon position for each coding gene are treated separately). To evaluate nodal support of the fully partitioned dataset we conducted a rapid bootstrap analysis in RAxML using 1000 bootstrap replicates. The resulting maximum-likelihood tree indicates that Isleria is not closely related to Myrmotherula or Epinecrophylla , but instead is sister to Thamnomanes Cabanis ( Fig. 1 View FIGURE 1 ).
Using the same partition strategy followed in the likelihood analysis, we also performed a Bayesian analysis as implemented in Mr. Bayes 3.1.2 ( Huelsenbeck & Ronquist, 2001) on the Cipres Science Portal ( Miller et al., 2010). To determine the best nucleotide substitution model for each partition, we used PAUP* ( Swofford, 2003) to obtain likelihood values for the 24 substitution models featured in MrModeltest 2.3 (Nylander, 2004). Based on the comparison of the AIC scores for each partition, we determined that the GTR + Γ + I model was the best fit for all codon positions of ND2 and the second codon position of ND3. GTR + Γ provided the best fit for the third codon position of RAG1 and RAG2, the third codon position of ND3 and cytb, and the nuclear intron BF5. GTR + I was the best fit for the second codon position of cytb; SYM + Γ + I provided the best fit for the first codon position of cytb; SYM + Γ was the best fit for the first codon position of ND3; HKY + Γ + I was the best fit for the first codon position of RAG2; and HKY + I was the best model for the second codon position of RAG1 and RAG2 and the first codon position of RAG1. We ran our analysis using 4 runs, 4 chains, 2 x 10 7 generations with a sample frequency of 1000, a burn-in of 20%, and chain temperature of 1.75. The use of the “compare” function of AWTY online ( Wilgenbusch et al., 2004) informed us that in all four runs we obtained highly similar levels of convergence. In addition, through the “slide” command we were able to establish that subsamples of the chains were sampling trees in proportion to their posterior probability. These two tests suggest that our phylogenetic hypothesis via Bayesian methods was satisfactory. The topology of the phylogenetic tree is virtually identical to that of the tree obtained through maximum-likelihood and also supports the separation of Isleria from Myrmotherula or Epinecrophylla ( Fig 2 View FIGURE 2. 50 ).
Both analyses indicated that I. guttata and I. hauxwelli are sister species and that they are distantly related to Myrmotherula and Epinecrophylla . Isleria and the genus Thamnomanes form sister groups embedded within a larger clade that contains a variety of morphologically, ecologically, and behaviorally distinctive genera such as Sclateria Oberholser , Phaenostictus Ridgway , Dichrozona , Thamnophilus Vieillot , Herpsilochmus Cabanis , Dysithamnus Cabanis , Taraba Lesson , and others. This result confirms that previously recognized assemblages ( Zimmer & Isler, 2003) within Myrmotherula represent independent evolutionary units and are not necessarily closely related. In this case, it had already been suggested that I. guttata and I. hauxwelli not only constitute a different assemblage ( Ridgely & Tudor, 1994) but could represent a separate evolutionary unit because of differences in plumage, vocalizations, ecology, and foraging behavior ( Zimmer & Isler, 2003).
An alternative treatment for the species placed in Isleria would be to merge them into Thamnomanes , but morphological, behavioral, vocal, and ecological differences do not warrant such taxonomic treatment. First, species of Thamnomanes lack markings on the apexes of wing coverts, tertials, and tail, whereas such markings are conspicuous in both species of Isleria . Second, both species of Isleria are much smaller ( 8.5–12 g) than species of Thamnomanes ( 16–21 g; Zimmer & Isler, 2003). Third, all species of Thamnomanes are known to be active sentinel participants in mixed-species flocks that forage in the understory and midstory (mostly up to 8 m above ground), whereas species of Isleria usually forage away from mixed-flocks and mostly below 3 m ( Pearson, 1977; Munn & Terborgh, 1979; Schulenberg, 1983; Stotz, 1990; Rosenberg, 1993; Zimmer & Isler, 2003). Finally, loudsongs of species of Thamnomanes are long series of notes that fall in pitch at the end before dropping into a final rattle or raspy note, whereas Isleria loudsongs are simpler and composed of repetitive whistles that become shorter at the end of the song ( Isler & Whitney, 2002; Zimmer & Isler, 2003). This level of differentiation is similar to that observed between other sister genera in the Thamnophilidae , and supports the separation of Thamnomanes and Isleria . Therefore, our decision to create a new genus ( Isleria ) is supported by the non-monophyly of the genus Myrmotherula , morphological, behavioral, and ecological differences with Thamnomanes , and the lack of an available genus name for I. guttata and I. hauxwelli .
TABLE 1. Ingroup taxa used in this study with frozen tissue collection voucher number. Tissue collections: LSUMZ—Louisiana State University Museum of Natural Science, Baton Rouge; USNM—United States National Museum of Natural History, Smithsonian Institution, Washington; FMNH—Field Museum of Natural History, Chicago; UWBM—University of Washington Burke Museum, Seattle; AMNH—American Museum of Natural History, New York City; INPA—Instituto Nacional de Pesquisas da Amazônia, Manaus, Brazil.
| Species | Subspecies | Tissue No. | Locality |
|---|---|---|---|
| Cymbilaimus lineatus | intermedius | LSUMZ B-18168 | Bolivia: Santa Cruz |
| Mackenziaena leachii | monotypic | USNM B-5986 | Argentina: Misiones |
| Taraba major | melanurus | FMNH 321773 | Peru: Madre de Dios |
| Thamnophilus doliatus | radiatus | UWBM RTB390 | Bolivia: Santa Cruz |
| Megastictus margaritatus | monotypic | LSUMZ B-6836 | Peru: Loreto |
| Neoctantes niger | monotypic | FMNH 321806 | Peru: Cusco |
| Dysithamnus mentalis | emiliae | FMNH 392443 | Brazil: Pernambuco |
| Thamnomanes ardesiacus | nominate | LSUMZ B-6896 | Peru: Loreto |
| Thamnomanes saturninus | nominate | FMNH 389947 | Brazil: Rondônia |
| Thamnomanes caesius | glaucus | USNM B-9482 | Guyana: Barima-Waini |
| Thamnomanes schistogynus | nominate | LSUMZ B-992 | Bolivia: La Paz |
| Epinecrophylla haematonota | nominate | LSUMZ B-4579 | Peru: Loreto |
| Epinecrophylla erythrura | septentrionalis | LSUMZ B-5474 | Peru: San Martín |
| Myrmotherula brachyura 1 | monotypic | LSUMZ B-4889 | Peru: Loreto |
| Myrmotherula brachyura 2 | monotypic | LSUMZ B-20305 | Brazil: Amazonas |
| Myrmotherula multostriata | monotypic | LSUMZ B-12968 | Bolivia: Santa Cruz |
| Myrmotherula gularis 1 | monotypic | LSUMZ B-16938 | Brazil: São Paulo |
| Myrmotherula gularis 2 | monotypic | FMNH 330815 | Brazil: São Paulo |
| Myrmotherula axillaris 1 | luctuosa | FMNH 392444 | Brazil: Pernambuco |
| Myrmotherula axillaris 2 | nominate | LSUMZ B-55209 | Suriname: Sipaliwini |
| Myrmotherula menetriesii | nominate | LSUMZ B-9759 | Bolivia: Pando |
| Isleria hauxwelli 1 | nominate | LSUMZ B-10787 | Peru: Ucayali |
| Isleria hauxwelli 2 | clarior | FMNH 392044 | Brazil: Mato Grosso |
| Isleria hauxwelli 3 | clarior | LSUMZ B-36639 | Brazil: Rondõnia |
| Isleria hauxwelli 4 | suffusa | LSUMZ B-4270 | Peru: Loreto |
| Isleria hauxwelli 5 | hellmayri | FMNH 391378 | Brazil: Pará |
| Isleria guttata 1 | monotypic | USNM B-14413 | Guyana: East Berbice-Corentyne |
| Isleria guttata 2 | monotypic | FMNH 391375 | Brazil: Amapá |
| Isleria guttata 3 | monotypic | AMNH DOT-3042 | Venezuela: Amazonas |
| Isleria guttata 4 | monotypic | INPA A-1258 | Brazil: Amazonas |
| Isleria guttata 5 | monotypic | INPA A-1766 | Brazil: Roraima |
| Dichrozona cincta | monotypic | FMNH 391144 | Bolivia: La Paz |
| Herpsilochmus sticturus | monotypic | USNM B-5228 | Guyana: Cuyuni-Mazaruni |
| Terenura sharpei | monotypic | LSUMZ B-39086 | Bolivia: Cochabamba |
| Cercomacra tyrannina | nominate | LSUMZ B-2273 | Panama: Darién |
| Sclateria naevia | nominate | FMNH 391418 | Brazil: Amapá |
| Myrmeciza pelzelni | monotypic | LSUMZ B-7523 | Venezuela: Amazonas |
| Myrmornis torquata | nominate | FMNH 389880 | Brazil: Rondõnia |
| Phaenostictus mcleannani | nominate | LSUMZ B-2135 | Panama: Darién |
No known copyright restrictions apply. See Agosti, D., Egloff, W., 2009. Taxonomic information exchange and copyright: the Plazi approach. BMC Research Notes 2009, 2:53 for further explanation.
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |