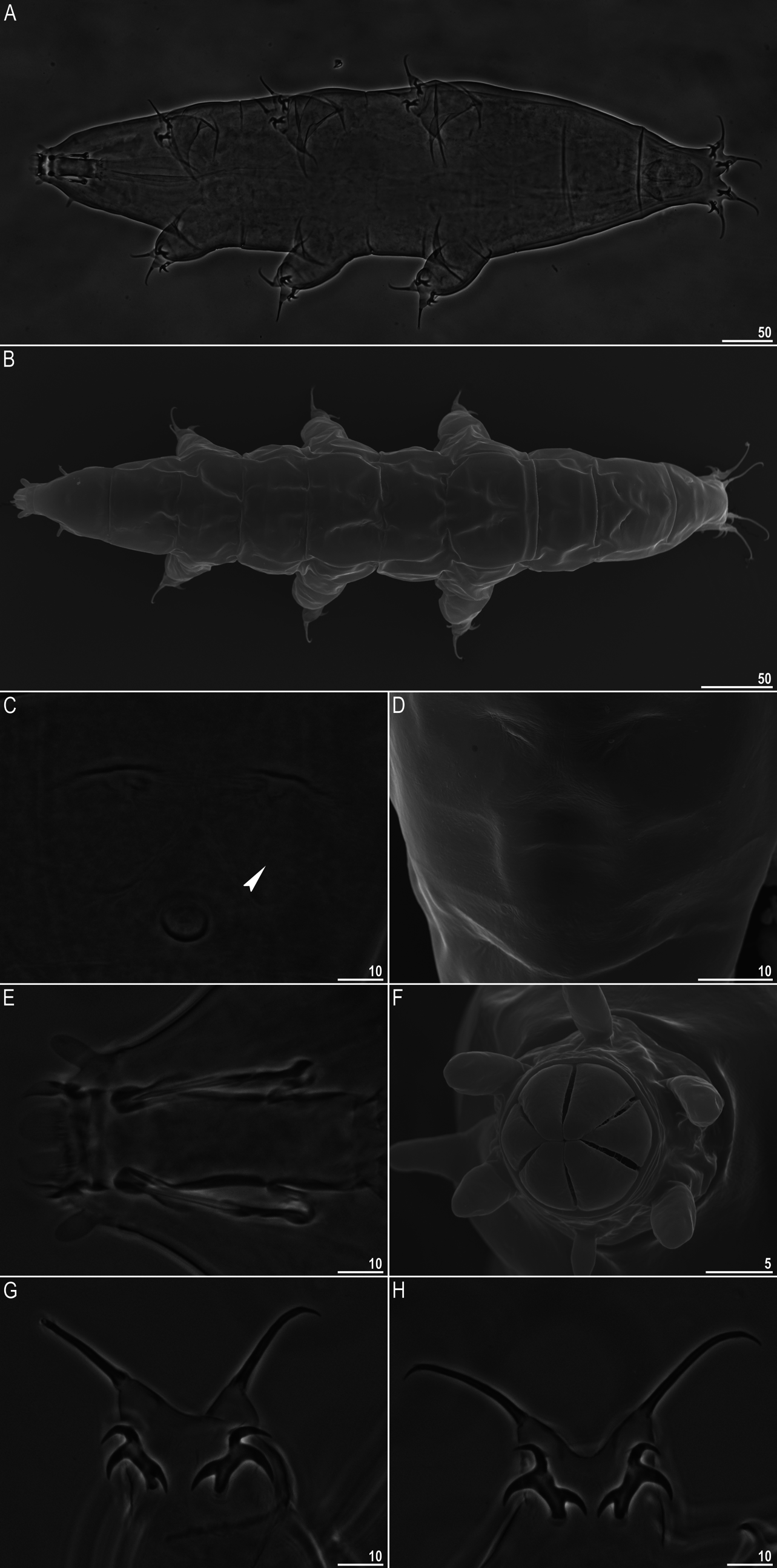
Milnesium inceptum, Morek & Suzuki & Schill & Georgiev & Yankova & Marley & Michalczyk, 2019
|
publication ID |
https://doi.org/10.11646/zootaxa.4586.1.2 |
|
publication LSID |
lsid:zoobank.org:pub:2D01FAB0-53DF-4B89-A891-A51C548D4B72 |
|
persistent identifier |
https://treatment.plazi.org/id/922D878E-DA47-3A78-FF14-FB4DFAFF161F |
|
treatment provided by |
Plazi |
|
scientific name |
Milnesium inceptum |
| status |
sp. nov. |
Delineation of M. alpigenum and M. inceptum sp. nov.
The two species are genetically distinct but morphologically very similar, although not identical. Therefore, they could be classified as pseudocryptic species, i.e. species that can be differentiated morphologically but only with a detailed analysis; in this case—with the use of statistical testing of morphometric traits measured in a number of specimens, since the classical identification based on qualitative traits and non-overlapping morphometric ranges is not sufficient to tell the species apart.
The PCA analysis indicated three components comprising 59.3% of the total variance in the pt ratios. The three factors were as follows: PC1: the pt of the primary and secondary branches of all claws (38.2%), PC2: the pt of external and posterior spurs (11.3%), and PC3: the pt of buccal tube widths and stylet support insertion point (9.8%). The relationships between the principal components are shown in Fig. 3 View FIGURE 3 . The two species did not differ in PC1 and PC3 but in contrast, they differed in PC2, thus comparisons of the first three principal components did not result in congruent conclusions. Specifically, when PC1 and PC3 were compared ( Fig. 3B View FIGURE 3 ), ranges for the two species largely overlapped. In the PC1 vs PC2 comparison ( Fig. 3A View FIGURE 3 ), the overlap between species was smaller. Finally, when PC2 and PC3 were compared, the ranges barely overlapped ( Fig. 3C View FIGURE 3 ). Thus, we compared only the traits constituting PC2 ( i.e. the pt values of claw spurs) with a series of t -tests and adjusted α-levels. Student’s t - tests revealed significant differences in three of the eight compared traits (mean values, ± SD, and [ranges] for M. alpigenum vs M. inceptum sp. nov.):
• spur on external claw I: 13.3 ± 1.5 [ 11.3–17.8] vs 11.3 ±1.3 [ 7.6–14.8], t 46 =4.002, p <0.001;
• spur on external claw II: 15.3 ±1.4 [ 12.7–17.5] vs 12.9 ±2.0 [ 9.2–16.6], t 54 =4.630, p <0.001;
• spur on external claw III: 15.2 ±1.4 [ 12.4–17.8] vs 12.4 ±1.8 [ 8.9–16.8], t 48 =5.758, p <0.001.
In other words, the analysis showed that M. alpigenum has statistically longer (relatively to the buccal tube length) external spurs than M. inceptum sp. nov. (compare also Fig. 1 View FIGURE 1 G–H and 2H–I).
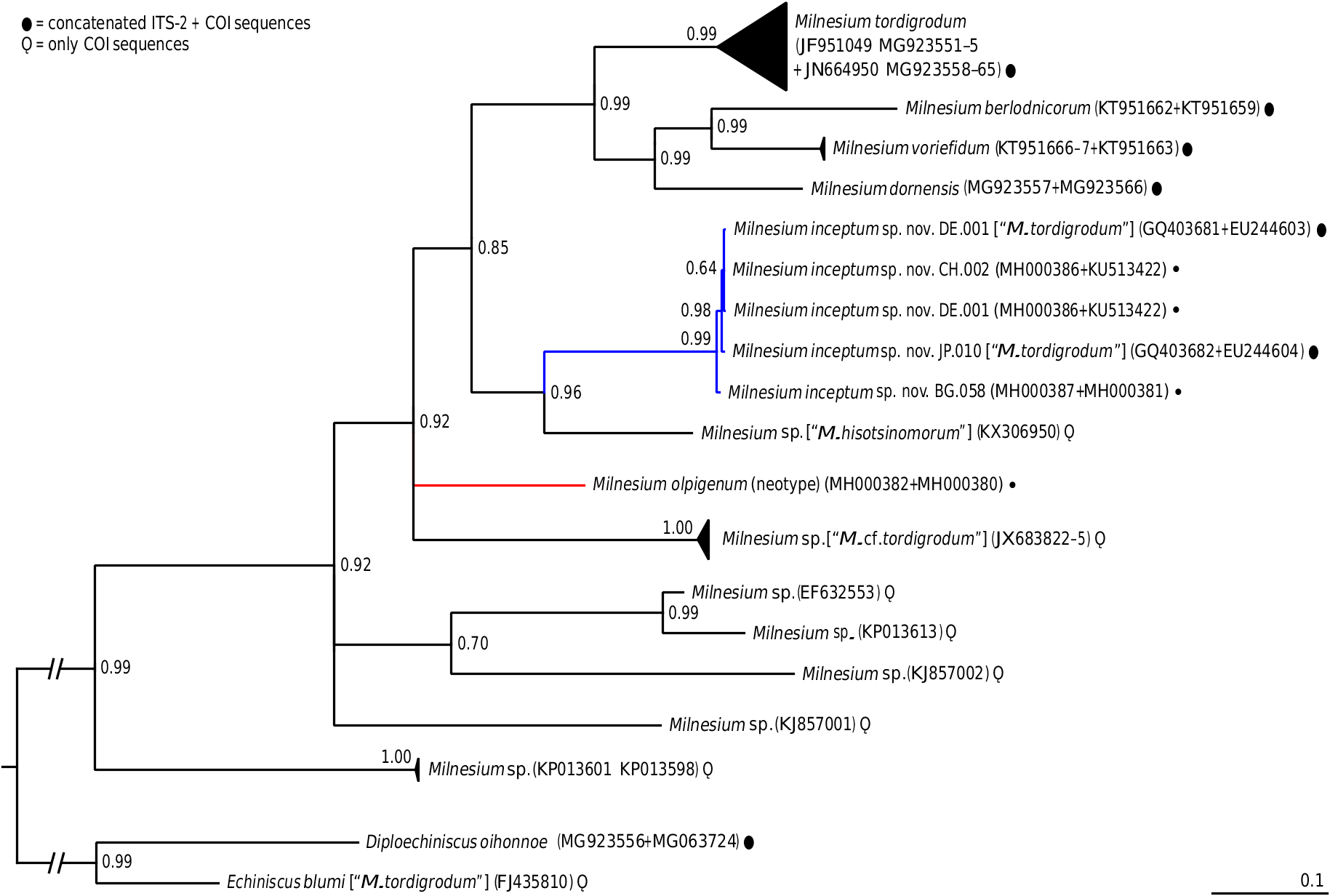
In contrast to subtle morphometric differences, the two species exhibit considerable genetic distances in all four analysed markers. Specifically, they differ by 1.0% in 18S rRNA, 5.2% in 28S rRNA, 21.6% in ITS-2, and by 18.1% in COI. Most importantly, however, the two species are not immediately related to each other (see Fig. 4 View FIGURE 4 for the positions of both species on the Milnesium phylogenetic tree), which is the strongest evidence that the two species represent separate phylogenetic lineages.
To conclude, differences both in phenotypic and genetic traits unequivocally show that M. inceptum sp. nov. is a bona species. Nevertheless, an extreme care must be taken when identifying these species using solely phenotypic data.
A B C 0.2 0.2
0.2
0.1 0.1 0.1
))) % % % 9 0 0... 2 0 0 1 0.0 1 0.0 1 0.0 (((2 3 3 PC PC PC
-0.1 -0.1 -0.1
-0.2
-0.2 -0.2
-0.2 0.0 0.2 -0.2 0.0 0.2 -0.1 0.0 -0.1 PC1 (3 5.8%) PC1 (3 5.8%) PC2 (1 2.9%)
No known copyright restrictions apply. See Agosti, D., Egloff, W., 2009. Taxonomic information exchange and copyright: the Plazi approach. BMC Research Notes 2009, 2:53 for further explanation.
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |